The STAT-SH2 Domain: Master Regulator of Dimerization in Health and Disease
This article provides a comprehensive analysis of the critical role played by the Src Homology 2 (SH2) domain in Signal Transducer and Activator of Transcription (STAT) protein dimerization—a pivotal event...
The STAT-SH2 Domain: Master Regulator of Dimerization in Health and Disease
Abstract
This article provides a comprehensive analysis of the critical role played by the Src Homology 2 (SH2) domain in Signal Transducer and Activator of Transcription (STAT) protein dimerization—a pivotal event in cellular signaling. Tailored for researchers and drug development professionals, we explore the unique structural features of STAT-type SH2 domains that facilitate both canonical phosphotyrosine-mediated dimerization and non-canonical interactions. The scope extends from foundational mechanisms and advanced research methodologies to the functional consequences of disease-associated mutations and emerging therapeutic strategies targeting the SH2 domain. By integrating foundational knowledge with current research and clinical implications, this review serves as a valuable resource for understanding and manipulating this crucial signaling node in cancer, immunology, and beyond.
The Structural Blueprint: Understanding the STAT-SH2 Domain Architecture
The Src Homology 2 (SH2) domain is a modular protein unit of approximately 100 amino acids that functions as a critical reader in phosphotyrosine-based signal transduction [1] [2]. Its primary function is to mediate specific protein-protein interactions by recognizing and binding to phosphorylated tyrosine (pTyr) containing peptide sequences on partner proteins [3]. This ability allows SH2 domain-containing proteins to participate in the assembly of multiprotein signaling complexes immediately downstream of protein tyrosine kinases (PTKs), thereby determining the specificity of cellular signaling pathways [3] [2]. In the context of STAT (Signal Transducer and Activator of Transcription) proteins, the SH2 domain plays an indispensable role in their activation cycle. It facilitates recruitment to activated cytokine receptors, mediates phosphorylation-dependent dimerization through reciprocal pTyr-SH2 domain interactions, and is essential for the nuclear translocation and transcriptional activity of STAT dimers [4] [5] [6]. Understanding the canonical structure of the SH2 domain, particularly its core αβββα motif and key sub-pockets, is therefore foundational to research focused on targeting STAT dimerization for therapeutic purposes.
The Canonical SH2 Domain Architecture
The Conserved αβββα Structural Core
All SH2 domains share a highly conserved tertiary structure despite significant sequence variation. The centerpiece of this structure is the αβββα motif, which forms a compact molecular scaffold [1] [7] [8]. This core fold consists of a central anti-parallel β-sheet (composed of three β-strands conventionally labeled βB, βC, and βD) that is flanked on both sides by two α-helices (αA and αB) [1] [7]. This arrangement creates a "sandwich"-like structure where the β-sheet partitions the ligand-binding surface of the domain into two functionally distinct sub-pockets [1]. The remarkable conservation of this three-dimensional fold across diverse proteins underscores its evolutionary optimization for recognizing phosphotyrosine motifs [8].
Generic Residue Numbering and Structural Nomenclature
To enhance comparability across different SH2 domains, a generic residue numbering scheme has been developed, inspired by similar systems in other protein families like GPCRs [1] [7]. In this scheme:
- Secondary structural elements are denoted by lowercase letters (a for α-helix, b for β-strand) with capital letters indicating their order in the N- to C-terminal direction [1].
- Within each structural element, the most conserved residue is assigned position 50 [1] [7].
- Residue numbers increase in the C-terminal direction and decrease in the N-terminal direction from this central point [1].
- Loops are named by combining the labels of the structural elements they connect (e.g., loop aAbB connects α-helix A to β-strand B) [1].
This nomenclature provides a unified framework for discussing and comparing structural features across the diverse SH2 domain family, which includes 120 distinct domains in the human proteome [7] [8].
Key Functional Sub-pockets and Their Molecular Anatomy
The central β-sheet strategically divides the SH2 domain's binding surface into two primary sub-pockets that jointly determine phosphopeptide recognition [1] [7].
Table 1: Key Functional Sub-pockets of the Canonical SH2 Domain
| Sub-pocket | Structural Components | Key Conserved Residues | Functional Role |
|---|---|---|---|
| Phosphotyrosine (pY) Pocket | αA helix, BC loop, one face of central β-sheet [5] | Sheinerman residues, including invariant Arg βB5 [1] [7] [8] | Anchors phosphorylated tyrosine via electrostatic interactions with phosphate moiety [1] [2] |
| Specificity (pY+3) Pocket | Opposite face of β-sheet, αB helix, CD and BC* loops [5] | Variable residues defining pocket chemistry and accessibility [9] [5] | Recognizes 3-5 amino acids C-terminal to pTyr, conferring binding specificity [9] [2] |
The Phosphotyrosine (pY) Binding Pocket
The pY pocket is a deeply buried, positively charged cavity that serves as the primary anchor for ligand binding. Its architecture features several highly conserved residues, most notably an invariant arginine at position βB5 (Arg175 in v-Src), which forms a critical salt bridge with the phosphate moiety of the phosphotyrosine [1] [2] [8]. This arginine is part of the FLXRXS signature motif (also known as the FLVR motif) found in the βB strand of nearly all SH2 domains [1] [7]. A group of eight conserved residues, collectively termed the Sheinerman residues, create the precise electrostatic environment necessary for high-affinity phosphate binding [1] [7]. The exceptional conservation of these residues highlights their fundamental role in the SH2 domain's function as a phosphorylation-dependent switch.
The Specificity (pY+3) Pocket
The pY+3 pocket, also referred to as the specificity-determining region, is more variable in sequence and structure across different SH2 domains [9] [5]. This pocket is responsible for recognizing specific amino acids at the +1 to +5 positions C-terminal to the phosphotyrosine, thereby conferring selectivity for distinct physiological ligands [3] [2]. The EF and BG loops play a particularly important role in defining the accessibility and shape of this pocket by either plugging or exposing key binding subsites [9]. For instance, in Grb2 SH2, a bulky tryptophan in the EF loop occupies the P+3 binding pocket, forcing the bound peptide to adopt a β-turn conformation and selecting for asparagine at the P+2 position [9] [2]. This loop-controlled access to binding pockets represents a fundamental mechanism for generating functional diversity within the conserved SH2 structural fold.
STAT-Type SH2 Domains: Specialization for Dimerization
STAT proteins feature a distinct subclass of SH2 domains that are structurally specialized for their unique role in transcription factor activation. STAT-type SH2 domains exhibit several characteristic features that distinguish them from Src-type SH2 domains [5] [8]:
- C-terminal Architecture: Instead of the βE and βF strands found in Src-type SH2 domains, STAT-type domains possess an additional α-helix (αB') in what is known as the evolutionary active region (EAR) [5] [8].
- Dimerization Interface: Residues in the αB, αB', and BC* loop participate in SH2-mediated STAT dimerization, forming critical cross-domain interactions that stabilize the parallel dimer configuration required for nuclear translocation [5].
- Flexibility: STAT SH2 domains exhibit significant structural flexibility, particularly in the pY pocket, which may facilitate their dual functions in phosphopeptide binding and dimer stabilization [5].
Table 2: Comparative Features of STAT-type versus Src-type SH2 Domains
| Feature | STAT-type SH2 Domains | Src-type SH2 Domains |
|---|---|---|
| C-terminal Structure | Additional α-helix (αB') [5] [8] | βE and βF strands with adjoining loop [8] |
| Primary Function | Receptor recruitment & STAT dimerization [5] | Signal relay & complex assembly [2] |
| Dimerization Role | Direct participation in parallel dimer formation [5] | Typically intramolecular autoinhibition [2] |
| Loop Characteristics | Shorter CD loops [8] | Variable, often longer loops [8] |
The critical importance of the STAT3 SH2 domain is highlighted by the numerous disease-associated mutations identified in patient sequencing studies. These mutations cluster in functionally significant regions and can result in either hyperactivated or loss-of-function STAT3 variants, underscoring the delicate balance required for proper SH2 domain function [5].
Experimental Approaches for Studying SH2 Domain Structure and Function
Methodologies for Mapping SH2 Domain Specificity
Understanding SH2 domain function requires precise characterization of their binding specificity and affinity. Several well-established experimental approaches are routinely employed:
Fluorescence Polarization (FP) Assays: This solution-based technique measures the change in polarization of fluorescently labeled phosphopeptides upon binding to SH2 domains. It provides quantitative data on binding affinity (Kd) and is particularly useful for competitive inhibition studies, such as those used to characterize STAT3 SH2 domain inhibitors [6]. The assay involves incubating purified SH2 domains with a fixed concentration of fluorescent tracer peptide and measuring polarization values across a range of protein concentrations or in the presence of potential inhibitors [6].
SPOT Peptide Array Analysis: This semiquantitative method involves synthesizing arrays of phosphorylated peptides on nitrocellulose membranes and probing them with recombinant SH2 domains [3]. The relative binding intensity to each peptide spot provides information about sequence preferences. This technique was instrumental in identifying the contextual dependence of SH2 domain recognition, including the role of both permissive and non-permissive residues in determining binding selectivity [3].
Oriented Peptide Array Library (OPAL) Screening: This approach uses degenerate phosphopeptide libraries to comprehensively map the binding preferences of SH2 domains [9]. The resulting specificity profiles have enabled the classification of SH2 domains into groups based on their preference for specific residues at the P+2, P+3, or P+4 positions C-terminal to the phosphotyrosine [9].
Structural Biology Techniques
- X-ray Crystallography: This remains the gold standard for determining high-resolution structures of SH2 domains in complex with phosphopeptide ligands. It has revealed diverse binding modes, including the classic two-pronged plug-and-socket model observed in Src family SH2 domains and the β-turn conformation seen in Grb2 SH2-peptide complexes [2].
- Nuclear Magnetic Resonance (NMR) Spectroscopy: NMR provides insights into the dynamics and conformational flexibility of SH2 domains in solution, complementing the static snapshots provided by crystallography [5].
Diagram 1: Experimental workflow for SH2 domain research, from initial database mining to therapeutic development.
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 3: Key Research Reagents and Experimental Tools for SH2 Domain Studies
| Reagent/Tool | Function/Application | Example Use Cases |
|---|---|---|
| GST-tagged SH2 Domains | Recombinant protein production for binding assays [3] | Purification via glutathione-Sepharose; FP and SPOT assays [3] |
| Phosphotyrosine Peptide Libraries | Mapping binding specificity and motifs [3] [9] | OPAL screening; SPOT array synthesis; competitive FP [3] [6] |
| STATeLight Biosensors | Real-time monitoring of STAT activation in live cells [10] | FLIM-FRET detection of STAT conformational changes [10] |
| SH2 Domain Inhibitors | Probing function and therapeutic development [6] | S3I-201, delavatine A derivatives for STAT3 dimer disruption [6] |
| Structural Biology Resources | Molecular visualization and analysis [1] [7] | SH2db database; PDB structures; AlphaFold models [1] [7] |
The canonical αβββα structure of the SH2 domain, with its bipartite organization into pY and pY+3 sub-pockets, represents an elegant evolutionary solution for achieving specific, phosphorylation-dependent protein-protein interactions. In STAT proteins, this conserved architecture has been specially adapted to facilitate the reciprocal SH2-phosphotyrosine interactions that underlie STAT dimerization and activation. The structural insights into these domains provide a crucial foundation for ongoing drug discovery efforts aimed at modulating STAT signaling in cancer and other diseases. Particularly promising are approaches that target the STAT3 SH2 domain to prevent dimerization, as demonstrated by inhibitors like S3I-201 and the delavatine A derivatives 323-1 and 323-2, which directly bind the SH2 domain and disrupt STAT3 dimer formation [6]. As structural databases like SH2db continue to expand and new methodologies such as genetically encoded biosensors enable real-time monitoring of STAT activation in live cells [10], our understanding of STAT-SH2 domain structure and function will continue to deepen, opening new avenues for therapeutic intervention in STAT-driven pathologies.
The Src Homology 2 (SH2) domain is a approximately 100-amino-acid modular unit that specifically recognizes and binds to phosphorylated tyrosine (pY) motifs, serving as a critical component in intracellular signal transduction [11] [12] [2]. Within the human proteome, SH2 domains are found in roughly 110 proteins involved in diverse cellular functions, including enzymes, adapters, and transcription factors [11] [8]. Despite a conserved core function of pY-recognition, SH2 domains have evolved structural variations that define their specific biological roles. STAT-type SH2 domains, found in Signal Transducers and Activators of Transcription (STAT) proteins, exhibit distinctive structural features that set them apart from the more common Src-type SH2 domains [5]. These differences are not merely structural curiosities; they are fundamental adaptations that enable the unique dimerization-dependent transcriptional functions of STAT proteins. Within the context of STAT dimerization research, understanding these structural distinctions is paramount, as the STAT SH2 domain is indispensable for mediating the reciprocal phosphotyrosine-SH2 interactions that form active dimers capable of nuclear translocation and DNA binding [4] [5]. This review delineates the unique structural, functional, and biophysical characteristics of STAT-type SH2 domains, contrasting them with Src-type domains, and explores the implications for experimental research and therapeutic intervention.
Structural Comparison: STAT-type vs. Src-type SH2 Domains
Consensus Fold and Fundamental Architecture
All SH2 domains share a conserved central αβββα structural motif, comprising a central anti-parallel β-sheet (βB-βD) flanked by two α-helices (αA and αB) [5]. This core scaffold creates two primary ligand-binding subpockets: the phosphotyrosine (pY) pocket, which binds the phosphate group of the phosphorylated tyrosine, and the pY+3 pocket, which confers binding specificity by interacting with residues C-terminal to the pY [5]. The pY pocket is highly conserved and features an invariant arginine residue (at position βB5) that forms a critical salt bridge with the phosphate moiety [11] [8]. Despite this common blueprint, STAT-type and Src-type SH2 domains diverge significantly in their C-terminal architectures and loop configurations, which directly influence their dimerization functions and ligand selectivity.
Table 1: Core Structural Features of SH2 Domain Types
| Feature | STAT-type SH2 Domains | Src-type SH2 Domains |
|---|---|---|
| C-terminal Structure | Features additional αB' helix [8] [5] | Contains βE and βF strands forming a small β-sheet [8] [5] |
| BC* Loop | Present; involved in dimerization interfaces [5] | Present; role primarily in ligand binding |
| Representative Proteins | STAT1, STAT3, STAT5 [5] | Src, Abl, Lck, Fyn [2] |
| Primary Functional Role | Mediate STAT dimerization for nuclear translocation and transcription [5] | Recruit signaling proteins to membranes and receptors; regulate catalytic activity [2] |
Determinants of Specificity and Binding
The specificity of SH2 domains is governed by interactions with amino acids flanking the phosphotyrosine, typically positions C-terminal to the pY [2]. While Src-type domains often recognize specific motifs extending from the pY (e.g., the Src SH2 domain prefers pYEEI), STAT-type SH2 domains are optimized for a unique function: they directly bind to phosphotyrosine motifs on other STAT monomers during activation-induced dimerization [5]. This reciprocal interaction creates a stable dimer that is essential for STAT function. The pY+3 pocket in STAT SH2 domains is therefore a critical determinant of dimerization specificity. Furthermore, the architecture of the STAT SH2 domain, particularly the presence of the αB' helix and the configuration of the BC* loop, creates a surface that facilitates specific cross-domain interactions between two STAT monomers, a feature not required in most Src-type domains [5].
Functional Implications for STAT Dimerization and Signaling
The unique structural features of STAT-type SH2 domains are direct adaptations for their non-redundant role in JAK-STAT signaling pathways. Upon activation by extracellular cytokines or growth factors, receptor-associated Janus Kinases (JAKs) phosphorylate specific tyrosine residues on the receptor's cytoplasmic tail. This creates docking sites for STAT proteins, which are subsequently recruited via their SH2 domains. Following recruitment, JAKs phosphorylate a single conserved tyrosine residue in the C-terminal transactivation domain of the STAT protein. This phosphorylation event triggers a profound conformational change: latent STAT monomers form active dimers via reciprocal phosphotyrosine-SH2 domain interactions [4] [5].
In this dimeric complex, the SH2 domain of one STAT monomer binds the phosphorylated tyrosine of its partner, and vice versa. The distinct structure of the STAT SH2 domain, especially the αB' helix and its surrounding regions, is crucial for stabilizing this dimeric interface. Once formed, the phosphorylated STAT dimer translocates to the nucleus, where it binds to gamma-activated sequence (GAS) elements in the promoters of target genes to regulate transcription [4] [5]. This entire process is dependent on the precise molecular architecture of the STAT SH2 domain. Disease-causing mutations frequently localize to this domain, disrupting dimerization and nuclear translocation, which underscores its functional importance [5]. For instance, germline heterozygous loss-of-function mutations in the STAT3 SH2 domain cause Autosomal-Dominant Hyper IgE Syndrome (AD-HIES), characterized by impaired Th17 T-cell responses [5].
The following diagram illustrates the central role of the SH2 domain in the canonical STAT activation and dimerization pathway.
Experimental Analysis of STAT-type SH2 Domains
Key Methodologies and Workflows
Deciphering the unique properties of STAT-type SH2 domains requires a multidisciplinary approach. The following experimental protocols are fundamental to probing their structure, dynamics, and function.
Protocol 1: Mapping SH2 Domain Specificity Using Peptide Array Libraries Objective: To define the phosphotyrosine-containing peptide sequence motif that a STAT SH2 domain recognizes with highest affinity [13].
- Clone and Express: Clone the cDNA encoding the STAT SH2 domain of interest into an expression vector. Express and purify the recombinant SH2 domain protein, often with an affinity tag (e.g., GST, His-tag) [13].
- Generate Peptide Library: Synthesize an oriented peptide array library (OPAL) on cellulose membranes. The library consists of thousands of spot-synthesized peptides, each containing a central phosphotyrosine flanked by degenerate amino acid sequences [13].
- Binding Assay: Incubate the purified SH2 domain with the peptide array membrane under controlled buffer conditions.
- Detection: Wash away unbound protein and detect the bound SH2 domain using a tag-specific antibody conjugated to a reporter enzyme (e.g., horseradish peroxidase) for chemiluminescent detection [13].
- Data Analysis: Identify the spots with the strongest signal intensity. Align the peptide sequences of the high-affinity binders to deduce the consensus binding motif (e.g., pY-X-X-Z, where X and Z are specific amino acids) [13].
Protocol 2: Characterizing Folding and Stability via Kinetic Analysis Objective: To determine the thermodynamic stability and folding pathway of a STAT SH2 domain, which can inform on the functional consequences of disease-associated mutations [12].
- Protein Purification: Purify the recombinant SH2 domain to homogeneity, ensuring it is suitable for biophysical analysis.
- Equilibrium Denaturation: Use a chemical denaturant like urea or guanidinium hydrochloride. Prepare a series of samples with increasing denaturant concentration. Monitor the unfolding transition using a spectroscopic signal intrinsic to the protein (e.g., fluorescence emission of tryptophan residues) or circular dichroism (CD) at 222 nm to track secondary structure loss [12].
- Stopped-Flow Kinetics: For folding/unfolding kinetics, use a stopped-flow instrument to rapidly mix the native or denatured protein with refolding or unfolding buffer, respectively. Monitor the change in fluorescence or CD signal over time (from milliseconds to seconds) [12].
- Data Modeling: Plot the observed rate constants against denaturant concentration to generate a "chevron plot." Analyze the plot to determine if folding follows a simple two-state model or involves populated intermediates. Extract thermodynamic parameters like the free energy of unfolding (ΔG) and the kinetic rate constants [12].
Protocol 3: Structural Workflow for SH2 Domain-Ligand Complexes Objective: To obtain high-resolution atomic-level structures of STAT SH2 domains, often in complex with phosphopeptide ligands, to visualize the dimerization interface and guide drug discovery [5].
- Crystallization: Crystallize the purified STAT SH2 domain, typically in the presence of a high-affinity phosphopeptide mimicking the binding partner. Optimization of crystallization conditions is critical.
- X-ray Diffraction: Flash-cool the crystal in liquid nitrogen and collect X-ray diffraction data at a synchrotron facility.
- Structure Determination: Solve the phase problem by molecular replacement using a known SH2 domain structure as a search model. Build and refine the atomic model into the electron density map.
- Analysis: Analyze the refined structure to identify key residues in the pY and pY+3 pockets, hydrogen bonding networks, and the conformational changes induced by peptide binding. Compare with known Src-type SH2 structures to highlight STAT-specific features [5].
The workflow for structural and biophysical characterization is summarized below.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Investigating STAT-type SH2 Domains
| Reagent / Tool | Function and Application | Technical Notes |
|---|---|---|
| Oriented Peptide Array Library (OPAL) | Defines the binding specificity and consensus motif of an SH2 domain [13]. | High-throughput; allows for screening of thousands of peptide sequences in parallel. |
| Phosphotyrosine Mimetic Peptides | Used in binding assays and crystallography to mimic the natural ligand and stabilize the SH2 domain structure [5]. | Often contain non-hydrolyzable pY mimetics like phosphonofluorophenylalanine (F2Pmp). |
| Site-Directed Mutagenesis Kits | Generates point mutations in the SH2 domain to probe the function of specific residues (e.g., in pY pocket) [12] [5]. | Critical for establishing structure-function relationships and validating disease mutations. |
| Recombinant SH2 Domain Proteins | Purified, often tagged (GST, His), proteins for in vitro binding, structural, and biophysical studies [13] [12]. | Essential for all downstream biochemical and structural analyses. |
| Denaturants (Urea, GdnHCl) | Used in equilibrium and kinetic folding experiments to unfold the SH2 domain and measure its stability [12]. | Allows for the determination of free energy of unfolding (ΔG) and identification of folding intermediates. |
Therapeutic Targeting and Research Perspectives
The STAT SH2 domain is a hotspot for mutations in diseases like cancer and immunodeficiencies, making it a prime target for therapeutic intervention [5]. Mutations can be either gain-of-function (GOF), leading to constitutive dimerization and oncogenic signaling (e.g., in T-cell leukemias), or loss-of-function (LOF), causing immunological deficiencies like AD-HIES [5]. The table below summarizes key disease-associated mutations.
Table 3: Exemplary Disease-Associated Mutations in the STAT3 SH2 Domain
| Mutation | Location/Region | Pathology | Functional Type | Molecular Consequence |
|---|---|---|---|---|
| S614R | BC Loop / pY Pocket | T-LGLL, NK-LGLL, ALCL [5] | Somatic GOF | Enhances phosphopeptide binding affinity, promoting constitutive activation. |
| E616K | BC Loop / pY Pocket | NKTL [5] | Somatic GOF | Disrupts autoinhibitory interactions, facilitating dimerization. |
| R609G | βB5 / pY Pocket | AD-HIES [5] | Germline LOF | Impairs phosphate coordination, crippling phosphotyrosine binding and dimerization. |
| S611I/N/G | βB7 / pY Pocket | AD-HIES [5] | Germline LOF | Disrupts the conserved SH2 fold, reducing stability and ligand binding. |
Targeting the STAT SH2 domain with small-molecule inhibitors presents a formidable challenge. The pY pocket is highly polar and charged, making it difficult to develop drug-like molecules with sufficient affinity and cell permeability [2] [5]. Furthermore, STAT SH2 domains exhibit significant conformational flexibility, with the pY pocket's accessible volume varying dramatically, complicating structure-based drug design [5]. Current strategies are exploring allosteric pockets, such as the evolutionary active region (EAR) in the pY+3 pocket, and developing non-peptidic, non-phosphorylated inhibitors that can disrupt the protein-protein interaction interface [11] [5]. The continued structural and biophysical dissection of STAT-type SH2 domains, particularly in the context of disease mutations, remains essential for unlocking their full potential as therapeutic targets.
The Src Homology 2 (SH2) domain serves as a fundamental modular component in intracellular signaling, specifically mediating protein-protein interactions through recognition of phosphorylated tyrosine (pTyr) residues. This in-depth technical guide examines the atomic-level mechanism of pTyr recognition by the conserved SH2 domain pocket, with particular emphasis on its indispensable role in STAT protein dimerization and signal transduction. We synthesize structural biology data, quantitative binding affinity measurements, and recent advances in specificity profiling to provide researchers and drug development professionals with a comprehensive framework for understanding this critical biological mechanism. The content further explores experimental methodologies for investigating SH2 domain interactions and discusses emerging therapeutic strategies targeting these interfaces.
Intracellular communication in metazoans relies heavily on tyrosine phosphorylation, a post-translational modification that creates docking sites for signaling proteins. The SH2 domain, a conserved protein module of approximately 100 amino acids, serves as the primary "reader" of these phosphotyrosine signals [14]. First identified in the v-Fps/Fes oncoprotein, SH2 domains have since been found in over 100 human proteins, including kinases, phosphatases, adaptor proteins, and transcription factors such as STATs (Signal Transducers and Activators of Transcription) [15]. These domains function as critical regulatory elements by directing the formation of transient protein complexes in response to tyrosine phosphorylation events. The specificity of SH2 domain-pTyr interactions ensures proper signal transmission from activated receptors to downstream pathways, including the canonical JAK-STAT signaling cascade [16]. In the context of STAT proteins, the SH2 domain performs a dual function: it mediates recruitment to phosphorylated receptor cytoplasmic tails and facilitates STAT dimerization through reciprocal SH2-pTyr interactions [17] [18]. This review comprehensively examines the structural mechanism of pTyr recognition by SH2 domains, with specific emphasis on its fundamental role in STAT biology and dimerization.
Structural Architecture of the SH2 Domain
Conserved SH2 Domain Fold
All SH2 domains adopt a conserved structural fold despite sequence variation, consisting of a central antiparallel β-sheet flanked by two α-helices [15]. The core structure comprises three or four β-strands forming the central sheet, surrounded by two α-helices (αA and αB) positioned on either side [16]. This conserved architecture creates a binding interface specifically designed for phosphopeptide recognition. The N-terminal region of the SH2 domain (from αA to βD) forms a highly conserved phosphotyrosine-binding pocket, while the C-terminal half (from βD to βG) exhibits greater structural variability that confers sequence specificity [14]. The most conserved residues cluster primarily on the βB strand, which contains the critical arginine residue responsible for coordinating the phosphate moiety of pTyr [14].
The Phosphotyrosine Binding Pocket
The pTyr binding pocket is a positively charged cleft on the SH2 domain surface that recognizes the phosphate group of phosphorylated tyrosine. A strictly conserved arginine residue (located within the highly conserved FLVR sequence motif on the βB strand) serves as the anchor point, forming bidentate hydrogen bonds with the phosphate moiety [15] [14]. This arginine-phosphate interaction provides approximately half of the total binding free energy for SH2 domain-phosphopeptide interactions [14]. The pocket is further defined by other positively charged and polar residues that stabilize the phosphate group through additional hydrogen bonding and electrostatic interactions. The depth and charge complementarity of this pocket explain the strong preference for phosphotyrosine over unmodified tyrosine or phosphoserine/threonine, ensuring specificity in signaling fidelity.
Table 1: Key Structural Elements of the SH2 Domain Fold
| Structural Element | Position in Fold | Functional Role | Conservation Level |
|---|---|---|---|
| Central β-sheet | Core domain | Provides structural scaffold | High |
| αA helix | N-terminal region | Flanks binding pocket | Medium-High |
| αB helix | C-terminal region | Contributes to specificity | Medium |
| βB strand | N-terminal half | Contains conserved Arg for pTyr binding | Very High |
| EF loop | Connects βE-βF | Determines specificity pocket access | Low (Variable) |
| BG loop | Connects βG-αB | Determines specificity pocket access | Low (Variable) |
Molecular Mechanism of pTyr Recognition
Canonical Binding Mode
In the canonical binding mode, phosphotyrosine-containing peptides bind to SH2 domains in an extended conformation, positioning perpendicular to the central β-strands of the domain [14]. The interaction involves two distinct binding pockets: (1) the conserved pTyr pocket that engages the phosphate group, and (2) a specificity pocket that recognizes residues C-terminal to the pTyr [16]. The pTyr residue inserts into the deep, basic pocket where the conserved arginine forms crucial hydrogen bonds with the phosphate group. The peptide backbone then extends across the SH2 domain surface, allowing the C-terminal flanking residues (typically positions +1 to +6 relative to pTyr) to interact with the specificity-determining region of the domain [16]. This two-pocket mechanism enables SH2 domains to achieve both high affinity (through phosphate coordination) and specificity (through interactions with flanking residues).
Specificity Determinants
SH2 domain specificity is primarily determined by interactions with amino acids at positions C-terminal to the phosphotyrosine residue. Structural studies reveal that the hydrophobic pocket located in the C-terminal half of the SH2 domain engages these flanking residues [14]. The EF and BG loops, which connect secondary structure elements, play a particularly important role in controlling access to this specificity pocket and thus dictate peptide selectivity [19] [14]. Different SH2 domains recognize distinct sequence motifs based on their unique complementarity-determining regions. For example, Src family kinases preferentially bind to pYEEI motifs, while the SH2 domains of PI3K or PLC-γ favor pYφXφ motifs (where φ represents a hydrophobic residue) [16]. This specificity allows different SH2 domain-containing proteins to engage distinct subsets of the phosphoproteome, enabling precise signaling pathway activation.
Table 2: SH2 Domain Specificity Profiles and Representative Binding Motifs
| SH2 Domain | Preferred Recognition Motif | Representative Binding Proteins | Affinity Range (K_D) |
|---|---|---|---|
| Src Family | pYEEI | Src, Fyn, Lck | 0.1-0.5 μM |
| Grb2 | pYXNX | Growth factor receptors | 0.2-5 μM |
| PI3K | pYφXM | PDGFR, IRS-1 | 0.5-5 μM |
| STAT1 | pYXPQ | IFN-γ receptor | 0.1-1 μM |
| ZAP70 | pYφXφ | T-cell receptor ζ-chain | 0.2-2 μM |
| PLC-γ | pYφXφ | PDGFR, FGFR | 0.3-3 μM |
SH2 Domains in STAT Dimerization and Activation
Dual Roles in STAT Signaling
STAT proteins exemplify the critical functional importance of SH2 domains in transcription factor regulation. The STAT SH2 domain performs two essential functions in JAK-STAT signaling: first, it mediates recruitment to phosphorylated cytokine receptors; second, it facilitates STAT dimerization through reciprocal SH2-pTyr interactions [17] [18]. In the canonical pathway, cytokine binding induces receptor dimerization and activation of associated JAK kinases, which phosphorylate tyrosine residues on the receptor cytoplasmic tails. STAT proteins are then recruited to these phosphotyrosine motifs via their SH2 domains [18]. Once bound, JAKs phosphorylate a conserved tyrosine residue in the STAT C-terminus, inducing a conformational change that enables STAT dimerization.
STAT Dimerization Mechanism
STAT dimerization occurs through reciprocal SH2-phosphotyrosine interactions between two STAT monomers [17]. Specifically, the SH2 domain of one STAT molecule binds to the phosphorylated tyrosine residue (pTyr) of its dimerization partner. This interaction forms stable STAT homo- or heterodimers that translocate to the nucleus and bind target DNA sequences [18]. Research on Stat1 and Stat2 has demonstrated that their SH2 domains mediate both homo- and heterodimerization, with evidence suggesting that a single SH2-phosphotyrosyl interaction is sufficient for dimer formation [17]. The crystal structures of STAT SH2 domains in complex with phosphopeptides reveal similar binding modes to other SH2 domains, with the conserved arginine coordinating the phosphate moiety and specificity pockets engaging residues C-terminal to the pTyr. This dimerization mechanism is conserved across STAT family members and is essential for their transcriptional activity.
Figure 1: STAT Activation Pathway via SH2-Mediated Dimerization. This diagram illustrates the sequential process from cytokine binding to STAT-mediated gene transcription, highlighting the critical SH2-pTyr interaction step.
Quantitative Analysis of SH2-pTyr Interactions
Binding Affinity and Kinetics
SH2 domain interactions with phosphotyrosine motifs are characterized by moderate affinity and specific kinetic parameters that enable dynamic signaling responses. Typical dissociation constants (K_D) for SH2 domain-phosphopeptide interactions range from 0.1 to 10 μM, with most physiological interactions falling between 0.2-5 μM [16] [14]. This moderate affinity is crucial for allowing transient association and dissociation events necessary for reversible signal transduction. Artificially increasing affinity through engineered "superbinder" SH2 domains has been shown to cause detrimental consequences to cells, demonstrating the physiological importance of this affinity range [14]. Kinetic studies reveal that SH2 domain interactions exhibit rapid association and dissociation rates, enabling responsive signaling systems. Comparative analysis with phosphotyrosine-binding (PTB) domains shows that PTB domain-mediated interactions can display similar overall affinity but slower exchange kinetics than SH2 domains, suggesting that PTB domains may not rapidly exchange among associated proteins [20].
Lipid Binding and Membrane Localization
Recent research has revealed an additional layer of complexity in SH2 domain function: approximately 90% of human SH2 domains can bind plasma membrane lipids independently of their phosphotyrosine recognition capability [21]. These lipid interactions occur through surface cationic patches separate from the pTyr-binding pocket, allowing simultaneous binding to both membranes and pTyr motifs [21]. The lipid-binding sites adopt two distinct configurations: grooves for specific phosphoinositide headgroup recognition or flat surfaces for non-specific membrane binding. Cellular studies with ZAP70 demonstrated that multiple lipids bind its C-terminal SH2 domain in a spatiotemporally specific manner, exerting exquisite control over its protein binding and signaling activities in T cells [21]. This dual-binding capability suggests that membrane localization may work in concert with pTyr recognition to enhance specificity and efficiency in signal transduction.
Experimental Methods for Profiling SH2-pTyr Interactions
High-Throughput Specificity Profiling
Advancements in peptide display technologies have enabled comprehensive profiling of SH2 domain specificity landscapes. A recently developed platform combines bacterial surface display of genetically encoded peptide libraries with deep sequencing to quantitatively analyze SH2 domain binding specificities [22]. This method involves displaying peptide libraries on the surface of E. coli cells as fusions to an engineered bacterial surface-display protein (eCPX). The libraries can include fully random sequences (X5-Y-X5 libraries) or defined sequences derived from human proteome phosphorylation sites (pTyr-Var libraries) [22]. SH2 domains of interest are used as baits to isolate binding peptides through magnetic bead-based separation, followed by deep sequencing of bound peptides to determine enrichment patterns. This approach allows for simultaneous processing of multiple samples and can profile specificity against libraries containing millions of peptide sequences, providing unprecedented resolution of SH2 domain recognition rules.
Figure 2: High-Throughput SH2 Specificity Profiling Workflow. This experimental pipeline illustrates the process from library generation to specificity analysis using bacterial peptide display and deep sequencing.
Structural and Biophysical Approaches
X-ray crystallography and NMR spectroscopy have been instrumental in elucidating the atomic-level details of SH2 domain-pTyr interactions. Crystallographic analyses of SH2 domain-phosphopeptide complexes have revealed the conserved binding mode and specific variations that account for differential specificity across the SH2 domain family [14]. NMR techniques provide complementary information about binding kinetics and dynamics under physiological conditions. Surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC) offer quantitative measurements of binding affinity and thermodynamics, enabling researchers to determine the energetic contributions of specific residues to the interaction [20]. These biophysical approaches are particularly valuable for characterizing the effects of disease-associated mutations and for evaluating potential therapeutic compounds that target SH2 domain interfaces.
Table 3: Experimental Methods for Studying SH2-pTyr Interactions
| Method | Key Output Parameters | Throughput | Information Gained |
|---|---|---|---|
| Bacterial Peptide Display | Specificity profiles, enrichment scores | High | Comprehensive recognition motifs |
| X-ray Crystallography | Atomic coordinates, binding interfaces | Low | Detailed molecular interactions |
| NMR Spectroscopy | Chemical shifts, dynamics, weak affinities | Medium | Solution-state structure and dynamics |
| Surface Plasmon Resonance | KD, kon, k_off | Medium | Binding kinetics and affinity |
| Isothermal Titration Calorimetry | K_D, ΔG, ΔH, ΔS | Low | Thermodynamic parameters |
| Oriented Peptide Libraries | Positional scanning data | Medium | Amino acid preferences at each position |
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Research Reagents for SH2 Domain Studies
| Reagent / Tool | Function/Application | Key Features | Example Uses |
|---|---|---|---|
| SH2 Domain Superbinders | High-affinity pTyr capture | Mutated pTyr pocket for enhanced affinity | Proteomic studies, detection assays |
| Phosphopeptide Libraries | Specificity profiling | X5-Y-X5 or proteome-derived sequences | High-throughput specificity screens |
| Bacterial Display Systems | Peptide library screening | eCPX-based display platform | SH2 ligand identification |
| Recombinant SH2 Domains | Biophysical characterization | Tagged purification (GST, His) | Structural studies, in vitro binding |
| JAK-STAT Reporter Cells | Functional signaling assays | STAT-responsive luciferase reporters | Pathway activation studies |
| Phosphospecific Antibodies | Detection of STAT phosphorylation | pY701-STAT1, pY705-STAT3 specific | Western blot, immunofluorescence |
The phosphotyrosine pocket of SH2 domains represents a remarkable example of evolutionary optimization for specific molecular recognition. The conserved structural framework coupled with variable specificity determinants enables these domains to mediate precise interactions in complex signaling networks. In STAT proteins, the SH2 domain plays an especially critical role in both receptor recruitment and dimerization, making it a central player in JAK-STAT signal transduction. Recent advances in understanding lipid binding by SH2 domains and high-throughput specificity profiling have expanded our appreciation of the regulatory complexity of these interactions. The experimental methodologies outlined here provide powerful approaches for investigating SH2 domain function and characterizing potential therapeutic interventions. As structural information grows and profiling technologies continue to advance, our understanding of the subtleties of SH2 domain specificity will further improve, facilitating the development of targeted therapies for diseases involving aberrant tyrosine kinase signaling, including cancer, immunodeficiencies, and inflammatory disorders.
The Specificity (pY+3) Pocket and Evolutionary Active Region (EAR)
The Src Homology 2 (SH2) domain is a critical modular unit found in numerous signaling proteins, including the Signal Transducer and Activator of Transcription (STAT) family. Its primary function is to recognize and bind phosphorylated tyrosine (pY) residues, thereby facilitating specific protein-protein interactions essential for cellular signaling [8]. Within the STAT protein structure, the SH2 domain serves a dual purpose: it mediates recruitment to activated cytokine receptors and is indispensable for the dimerization of STAT monomers through reciprocal phosphotyrosine-SH2 domain interactions [23]. This dimerization is a fundamental step in the canonical STAT activation pathway, enabling nuclear translocation and regulation of target genes.
This whitepaper focuses on two specialized structural regions within the STAT-SH2 domain: the specificity pocket (pY+3) and the Evolutionary Active Region (EAR). These regions are crucial for determining binding selectivity and facilitating the structural adaptations necessary for STAT dimerization. Understanding their precise mechanisms provides a foundation for targeted therapeutic interventions aimed at modulating pathological STAT signaling in cancer and inflammatory diseases [23].
Structural Anatomy of the STAT-SH2 Domain
The STAT-SH2 domain shares a conserved fold but possesses distinct features that differentiate it from other SH2 domains, such as those in the Src kinase family.
Core SH2 Domain Architecture
The fundamental structure of an SH2 domain consists of a central anti-parallel β-sheet (comprising strands βB, βC, and βD) flanked by two α-helices (αA and αB), forming an αβββα motif [23]. This core structure creates two primary ligand-binding pockets:
- The pY pocket: A deep pocket that binds the phosphate moiety of the phosphorylated tyrosine residue. It contains a highly conserved arginine residue (from the FLVR motif) that forms a salt bridge with the phosphotyrosine [8] [23].
- The pY+3 pocket: Located adjacent to the pY pocket, it accommodates the amino acid residue at the +3 position relative to the phosphotyrosine in the peptide ligand. This pocket is the primary determinant of binding specificity [23].
Unique Characteristics of the STAT-Type SH2 Domain
STAT-type SH2 domains are characterized by specific structural deviations from the Src-type [8] [24]:
- They lack the C-terminal βE and βF strands typically found in Src-type SH2 domains.
- The αB helix is split into two helices, designated as αB and αB'.
- The region encompassing the αB' helix and the adjacent BC* loop (connecting the αB and αC helices) constitutes the Evolutionary Active Region (EAR) [23]. This area is a hotspot for structural variation and disease-associated mutations.
Table 1: Key Structural Regions of the STAT-SH2 Domain
| Structural Region | Key Components | Primary Function |
|---|---|---|
| pY Pocket | αA helix, BC loop, βB strand, conserved Arg | High-affinity binding to phosphotyrosine |
| pY+3 Pocket | Opposite face of central β-sheet, αB helix, CD loop | Determines ligand-binding specificity |
| Evolutionary Active Region (EAR) | αB' helix, BC* loop | Facilitates STAT dimerization and cross-domain interactions |
The pY+3 Pocket: Determining Specificity
The pY+3 pocket is a specificity-determining cavity within the SH2 domain that engages residues C-terminal to the phosphotyrosine. Its chemical and physical properties dictate which peptide sequences the SH2 domain can recognize with high affinity.
Mechanism of Specificity Determination
The binding of a phosphopeptide to an SH2 domain occurs perpendicular to the central β-sheet. While the pY residue anchors the interaction, the amino acids at the +1, +2, and especially the +3 position relative to the pY extend into the pY+3 pocket. The physicochemical environment of this pocket—dictated by the side chains of the surrounding SH2 domain residues—selectively favors certain amino acids over others, enabling the discrimination between different STAT proteins and other SH2-containing signaling molecules [23]. This ensures the fidelity of downstream signaling cascades.
Role in STAT Dimerization
In the context of STAT activation, the pY+3 pocket is critical for reciprocal SH2-phosphotyrosine interaction that forms active STAT dimers. One STAT monomer presents its phosphorylated tyrosine, while the other monomer's SH2 domain binds to it, with the pY+3 pocket playing a key role in stabilizing this interaction. This dimerization is a prerequisite for the nuclear accumulation of STATs and their function as transcription factors [23].
The Evolutionary Active Region (EAR): A Hub for Regulation and Mutation
The Evolutionary Active Region is a defining feature of STAT-type SH2 domains and represents a critical functional and evolutionary module.
Structural Composition of the EAR
The EAR is located at the C-terminal end of the pY+3 pocket and includes two key elements:
- The αB' Helix: An additional α-helix not found in Src-type SH2 domains.
- The BC* Loop: The loop connecting the αB and αC helices [23].
This region participates in cross-domain interactions that stabilize the dimeric structure of activated, phosphorylated STATs. Its evolutionary conservation underscores its fundamental role in STAT protein function.
The EAR as a Mutational Hotspot
Sequencing of patient samples has identified the SH2 domain, and the EAR in particular, as a hotspot for disease-associated mutations [23]. These mutations can have either gain-of-function (GOF) or loss-of-function (LOF) consequences, disrupting the delicate balance of cellular signaling.
Table 2: Selected Disease-Associated Mutations in the STAT3/STAT5 SH2 Domain
| STAT Protein | Mutation | Location | Functional Impact | Associated Disease |
|---|---|---|---|---|
| STAT3 | Various point mutations | SH2 domain (incl. EAR) | Loss-of-Function (LOF) | Autosomal-Dominant Hyper IgE Syndrome (AD-HIES) [23] |
| STAT3 | Various point mutations | SH2 domain (incl. EAR) | Gain-of-Function (GOF) | Autoimmune disorders, lymphoproliferative disease [23] |
| STAT5B | Various point mutations | SH2 domain (incl. EAR) | Loss-of-Function (LOF) | Growth hormone insensitivity syndrome (GHIS) [23] |
The genetic volatility of the EAR highlights its evolutionary active nature. Even single amino acid changes can alter dimerization stability, DNA-binding affinity, and transcriptional output, making it a compelling target for drug discovery.
Experimental Analysis of the pY+3 Pocket and EAR
Investigating the structure and function of these regions requires a combination of biophysical, computational, and cell-based assays.
Structural Methodologies
X-ray Crystallography and NMR Spectroscopy: These high-resolution techniques are fundamental for determining the three-dimensional structures of STAT-SH2 domains, both alone and in complex with phosphopeptide ligands. They allow for the precise mapping of the pY+3 pocket and visualization of the EAR's role in dimer interface formation [23].
Isothermal Titration Calorimetry (ITC) and Surface Plasmon Resonance (SPR): These methods are used to quantitatively measure the binding affinity (Kd) and thermodynamics of interactions between SH2 domains and their target phosphopeptides. They are essential for characterizing how mutations in the pY+3 pocket or EAR affect ligand binding [8] [23].
Functional and Cellular Assays
Electrophoretic Mobility Shift Assay (EMSA): This assay assesses the DNA-binding capability of STAT dimers. Mutations that impair dimerization via the SH2 domain will result in reduced DNA binding activity [4].
Luciferase Reporter Assays: To functionally validate the impact of SH2 domain mutations, a luciferase gene under the control of a STAT-responsive promoter is transfected into cells. Changes in luciferase activity upon cytokine stimulation directly report on the transcriptional activity of the STAT pathway [23].
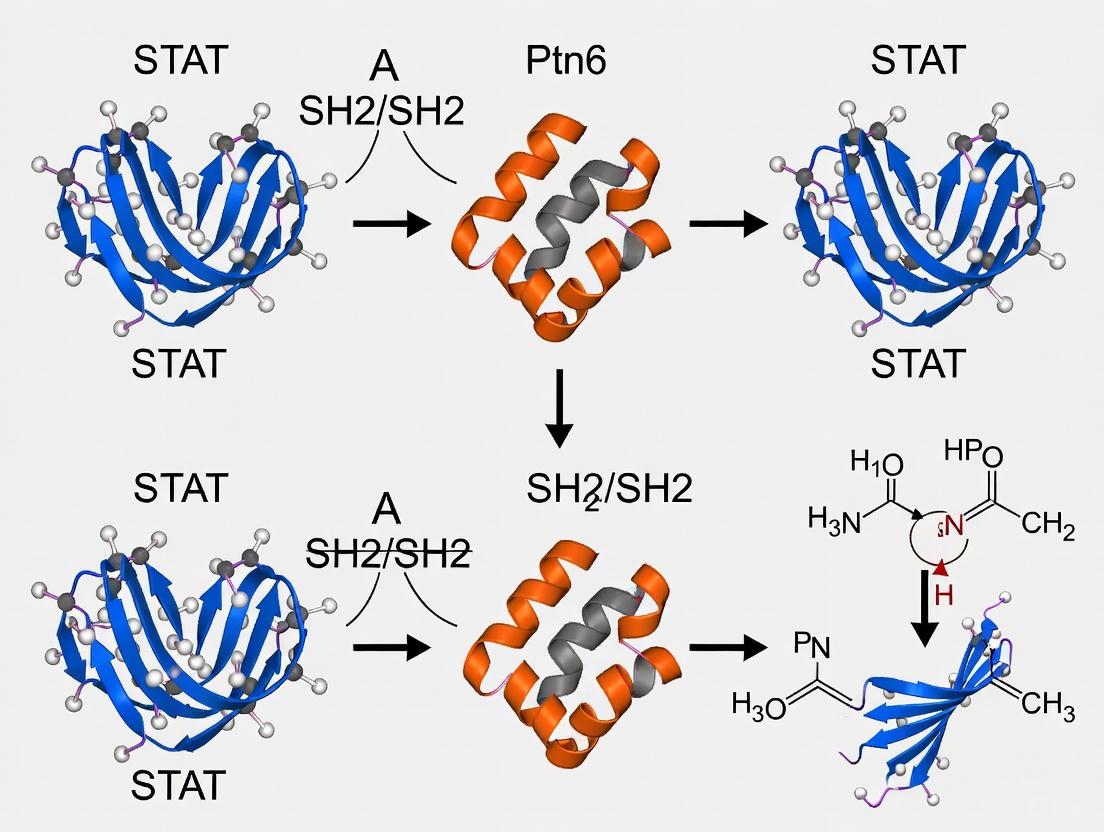
CoDIAC Analysis: A comprehensive computational approach for domain interface analysis. As detailed in a 2025 study, CoDIAC is a structure-based interface analysis tool that can map domain interfaces from experimental and predicted structures. It can identify conserved post-translational modifications (PTMs) relative to interaction interfaces, enabling researchers to infer the specific effects of mutations or PTMs on SH2 domain function and regulation [25].
The Scientist's Toolkit: Research Reagent Solutions
Progress in STAT-SH2 domain research relies on a suite of specialized reagents and tools.
Table 3: Essential Research Reagents for STAT-SH2 Domain Studies
| Reagent / Tool | Function & Application | Key Characteristics |
|---|---|---|
| Recombinant STAT-SH2 Proteins | In vitro binding assays, structural studies, inhibitor screening. | Wild-type and mutant variants; full-length or truncated domains. |
| Phosphopeptide Ligands | Mapping binding specificity, competition assays, SPR/ITC experiments. | Sequences derived from native STATs or receptors; pY at defined position. |
| Site-Directed Mutagenesis Kits | Introducing disease-associated or rational mutations into the pY+3 pocket or EAR. | Essential for establishing structure-function relationships. |
| STAT-Luciferase Reporter Constructs | Cellular functional assays to measure pathway activity. | Reporter gene under control of STAT-responsive promoter. |
| CoDIAC Python Package | Computational analysis of SH2 domain interfaces, interactions, and regulation. | Reveals coordinated regulation by PTMs; infers effects of mutations [25]. |
Therapeutic Targeting and Future Directions
The critical role of the STAT-SH2 domain in dimerization makes it a high-priority target for therapeutic intervention, particularly in cancers driven by constitutive STAT signaling.
Drug discovery efforts have focused on developing small-molecule inhibitors that disrupt the protein-protein interactions mediated by the SH2 domain. The pY+3 pocket and the adjacent EAR are attractive targets due to their role in determining specificity and dimerization. However, the flexibility of the STAT SH2 domain and the relatively shallow nature of its binding surfaces have posed significant challenges [23]. Emerging strategies include:
- Targeting allosteric sites that indirectly affect the pY+3 pocket.
- Developing stapled peptides that mimic the native phosphopeptide and competitively inhibit dimerization.
- Exploiting insights from the CoDIAC framework to understand how post-translational modifications like acetylation or serine phosphorylation might regulate SH2 domain function and offer new avenues for intervention [25].
The specificity (pY+3) pocket and the Evolutionary Active Region (EAR) are integral components of the STAT-SH2 domain, jointly governing phosphopeptide recognition selectivity and STAT dimerization stability. Their distinct structural features within the STAT family and their propensity for disease-driving mutations underscore their biological and clinical significance. Continued research using integrated structural, biophysical, and computational methods will be essential to fully elucidate their mechanisms and to translate this knowledge into novel therapeutics for STAT-driven diseases.
The Src Homology 2 (SH2) domain is a approximately 100-amino-acid protein module that serves as a crucial recognition domain in tyrosine kinase signaling pathways by specifically binding to phosphorylated tyrosine (pY) residues [26] [2]. These domains are fundamental components of eukaryotic cellular communication, enabling the assembly of multiprotein signaling complexes in response to tyrosine phosphorylation events [2] [8]. While the canonical function of SH2 domains involves phosphotyrosine recognition, emerging research highlights the critical importance of SH2 domain flexibility and structural dynamics in regulating their biological functions [27] [28] [29]. This review explores the structural plasticity of SH2 domains, with particular emphasis on the STAT-SH2 domain, examining how conformational dynamics influence dimerization mechanisms and create novel opportunities for therapeutic intervention in disease states, particularly cancer and immune disorders.
Structural Fundamentals of SH2 Domains
Conserved Architecture with Adaptive Features
SH2 domains maintain a highly conserved structural fold despite sequence diversity across family members [8]. The core structure consists of a central anti-parallel β-sheet flanked by two α-helices, forming a compact globular domain [2] [8]. This scaffold creates two functionally critical binding pockets: a highly conserved phosphotyrosine (pY) binding pocket that engages the phosphorylated tyrosine residue, and a specificity-determining pocket that recognizes residues C-terminal to the phosphotyrosine, typically at the pY+3 position [2] [8].
The N-terminal region of SH2 domains is particularly conserved, featuring a deep pocket within the βB strand that contains an invariant arginine residue (at position βB5) responsible for coordinating the phosphate moiety of phosphotyrosine [8]. In contrast, the C-terminal region displays greater variability, contributing to ligand specificity diversity among different SH2 domains [8]. Structural variations in loop regions between secondary elements, particularly the EF loop and BG loop, further modulate binding specificity by controlling access to ligand-binding pockets [8].
Table 1: Key Structural Elements of SH2 Domains and Their Functional Roles
| Structural Element | Location | Primary Function | Conservation |
|---|---|---|---|
| βB strand & FLVR motif | pY binding pocket | Phosphate group coordination via invariant arginine | High across family |
| Specificity pocket | Adjacent to pY site | Recognition of C-terminal residues (e.g., pY+3) | Variable |
| EF & BG loops | Surface accessibility | Control ligand access to binding pockets | Moderate to variable |
| CD loop | Distal surface | Allosteric regulation, interdomain communication | Variable |
| Central β-sheet | Structural core | Domain stability, conformational transmission | High |
STAT-SH2 Domain Distinctiveness
STAT (Signal Transducer and Activator of Transcription) proteins feature a distinct subclass of SH2 domains classified as the "STAT-type," which differs structurally from the "Src-type" SH2 domains [8]. STAT-SH2 domains lack the βE and βF strands present in Src-type domains and feature a split αB helix [8]. This structural adaptation appears specialized to facilitate the reciprocal phosphotyrosine-SH2 domain interactions that underlie STAT dimerization and nuclear translocation following activation [4] [30].
The STAT-SH2 domain is essential for the canonical activation mechanism of STAT proteins [4]. Upon tyrosine phosphorylation by Janus kinases (JAKs) or receptor tyrosine kinases, STAT proteins form parallel dimers through reciprocal phosphotyrosine-SH2 domain interactions between two monomers [4] [30]. These dimers then translocate to the nucleus and bind specific DNA elements to regulate target gene expression [4]. Recent evidence indicates that unphosphorylated STAT (U-STAT) proteins can also form dimers and regulate gene expression through distinct mechanisms, expanding the functional repertoire of STAT-SH2 domains beyond canonical signaling [4].
Flexibility and Dynamics of SH2 Domains
Allosteric Communication Networks
SH2 domains function not as static binding modules but as dynamic structures with internal allosteric communication networks. Research on the Fyn SH2 domain demonstrates that information transfer occurs between the phosphopeptide binding site and distal regions of the domain via a contiguous pathway of residues that crosses the protein core [27]. This communication channel enables binding events at one site to trigger conformational changes at distal locations, facilitating coordination with other protein domains.
Molecular dynamics simulations and mutual information analysis of the Fyn SH2 domain have quantified these allosteric networks, revealing that the SH2 domain forms a "noisy communication channel" that couples residues in the phosphopeptide binding site with residues near the linkers connecting the SH2 domain to SH3 and kinase domains [27]. This connectivity allows ligand binding to influence domain orientation and interdomain relationships, effectively transmitting biological information across the protein structure.
Figure 1: Allosteric Communication in SH2 Domains. Ligand binding triggers information flow through internal networks to distal functional sites.
Loop Dynamics and Functional Regulation
Surface loops connecting secondary structural elements serve as critical mediators of SH2 domain flexibility and function. The CD loop, in particular, exemplifies how structural dynamics can influence signaling outcomes. Comparative studies between Csk and Src SH2 domains reveal that natural variations in CD loop length and flexibility significantly impact protein dynamics and catalytic efficiency, despite preservation of the global domain fold [28].
In Csk, engineering a more flexible CD loop through insertion of two glycine residues (to mimic the longer loop found in Src) resulted in reduced catalytic activity without affecting global domain folding or phosphopeptide binding capability [28]. This demonstrates that subtle modifications to flexible loop regions can alter allosteric communication pathways and functional output. Molecular dynamics simulations identified specific signal transduction routes from the distal CD loop to the active site, underscoring how surface loops can serve as tunable modulators of SH2 domain function [28].
STAT-SH2 Domain in STAT Dimerization
Molecular Determinants of STAT Dimerization
The STAT-SH2 domain mediates dimerization through a sophisticated interface involving multiple interaction types. Molecular dynamics simulations of STAT5A have revealed three distinct interfaces in the active dimer [30]:
- Reciprocal intermolecular PTM-SH2 domain interactions: The classical phosphotyrosine-SH2 domain interaction present in all activated STAT dimers
- Intermolecular PTM-PTM interactions: Interactions between C-terminal tail segments of opposing monomers
- Intramolecular PTM-SH2 domain interactions: Interactions within individual monomers that stabilize the dimer configuration
The phosphorylated tyrosine (pY694 in STAT5A) forms salt bridges with the invariant arginine (R618) in the βB strand of the opposing monomer's SH2 domain [30]. This primary interaction is supplemented by additional contacts involving residues K600, S620, S622, and T628, which form transient hydrogen bonds with the phosphotyrosine [30]. STAT5-specific hydrophobic interactions further stabilize this interface, with residues N642 and K644 engaging the aromatic ring of phosphotyrosine, and V695 (pY+1) packing into a hydrophobic pocket containing W631, W641, and L643 [30].
Novel Intramolecular Interface in STAT5
A distinctive feature of STAT5 dimerization is the presence of a novel intramolecular interaction mediated by F706, located adjacent to the phosphotyrosine motif, and a unique hydrophobic interface on the SH2 domain surface [30]. This interaction is dispensable for receptor-mediated phosphorylation of STAT5 but essential for dimer formation and subsequent nuclear accumulation [30]. Mutational analysis confirms that disruption of this hydrophobic interface abolishes STAT5 dimerization without affecting phosphorylation, highlighting its specific role in stabilizing the dimeric state.
This intramolecular interface differs significantly from corresponding regions in STAT1 and STAT3, which utilize flexible loop structures to stabilize dimerization [30]. The absence of conservation in these structural elements between STAT family members suggests distinct evolutionary solutions to the challenge of stable dimer formation, with implications for selective therapeutic targeting.
Table 2: Key Interactions in STAT-SH2 Domain Dimerization
| Interaction Type | Key Residues | Functional Role | Conservation |
|---|---|---|---|
| Phosphotyrosine coordination | pY694, R618, K600, S620, S622, T628 | Primary dimer interface formation | High across STATs |
| Hydrophobic packing | V695, W631, W641, L643 | Stabilization of pY positioning | Moderate |
| STAT5-specific hydrophobic | N642, K644 | Enhanced pY binding stability | STAT5-specific |
| Intramolecular interface | F706, hydrophobic surface | Dimer stabilization post-phosphorylation | STAT5-specific |
| PTM-PTM interface | Q698, I699, K700, Q701, E705 | Intermonomer contacts in C-terminal tails | Variable |
Methodological Approaches for Studying SH2 Dynamics
Computational and Theoretical Frameworks
Molecular dynamics (MD) simulations have proven invaluable for characterizing SH2 domain flexibility and allosteric communication. MD simulations of STAT5A, spanning 2000 nanoseconds, have enabled detailed analysis of dimer interface stability and identification of key interacting residues [30]. Similarly, enhanced sampling MD combined with small-angle X-ray scattering (SAXS) has successfully mapped allosteric interfaces between SH2 and kinase domains in Btk [29].
Information theory approaches provide complementary insights into SH2 domain dynamics. By applying mutual information analysis to SH2 domains, researchers can quantify conformational correlations between residues and map information exchange pathways across the protein structure [27]. This method treats the SH2 domain as a communication channel, quantifying how binding-induced conformational changes propagate through the structure to distal functional sites.
Experimental Techniques for Dynamics Analysis
Nuclear magnetic resonance (NMR) spectroscopy offers powerful approaches for characterizing SH2 domain dynamics at atomic resolution. Backbone dynamics studies using 15N relaxation experiments can probe changes in molecular motions upon ligand binding [31]. NMR-based hydrogen-deuterium exchange (HDx) experiments provide site-specific information on protein stability and dynamics by monitoring the exchange rates of backbone amide protons [28].
Biophysical and biochemical approaches further illuminate SH2 domain function. Fluorescence polarization assays quantitatively measure phosphopeptide binding affinities [29], while circular dichroism spectroscopy and thermal shift assays assess domain folding and stability [29]. Enzyme kinetic assays complement these approaches by quantifying the functional consequences of structural perturbations on catalytic activity [28] [29].
Table 3: Experimental Methods for Analyzing SH2 Domain Flexibility and Function
| Method | Application | Key Information | References |
|---|---|---|---|
| Molecular Dynamics Simulations | Conformational sampling, allosteric pathways | Residue correlations, dynamic interfaces | [27] [30] [29] |
| Mutual Information Analysis | Information transfer mapping | Communication pathways, allosteric networks | [27] |
| NMR Relaxation | Backbone dynamics | Ps-ns timescale motions, binding effects | [31] |
| Hydrogen-Deuterium Exchange | Stability and dynamics | Structural flexibility, stabilization effects | [28] |
| Fluorescence Polarization | Binding affinity | Ligand binding constants, specificity | [29] |
| Circular Dichroism | Secondary structure | Folding state, stability | [29] |
| Enzyme Kinetics | Functional output | Catalytic efficiency, allosteric effects | [28] [29] |
Therapeutic Targeting of SH2 Domains
Allosteric Targeting Strategies
The critical role of SH2 domains in signaling pathways and their involvement in disease states makes them attractive therapeutic targets. Traditional approaches focused on developing phosphotyrosine mimetics to compete with native ligands for the pY binding pocket, but these faced challenges due to poor pharmacological properties [2] [8]. Emerging strategies now target allosteric sites and dynamic interfaces, offering potentially more selective interventions.
Research on Btk exemplifies the therapeutic potential of allosteric SH2 domain targeting. A engineered repebody protein that binds the Btk SH2 domain and disrupts its interface with the kinase domain effectively prevents Btk activation in cells, inhibiting signaling and proliferation in malignant B-cells [29]. This approach remains effective against Btk with the C481S mutation that confers resistance to covalent ATP-competitive inhibitors, highlighting the value of allosteric targeting for overcoming drug resistance [29].
Targeting STAT-SH2 Dimerization Interfaces
The STAT-SH2 dimerization interface represents a promising target for therapeutic intervention in cancers and immune disorders driven by constitutive STAT signaling. The identification of a novel intramolecular interface in STAT5, mediated by F706 and a unique hydrophobic surface on the SH2 domain, provides a potential target for disrupting STAT5 dimerization [30]. This interface is particularly significant as it is dispensable for phosphorylation but essential for dimer formation, offering opportunity for selective inhibition without affecting upstream signaling events.
Several leukemic STAT5 mutants map to the SH2 domain and C-terminal tail segment, including T628S, N642H, Y665F, I699L, and Q701L [30]. These mutations promote constitutive STAT5 activation through various mechanisms, often by enhancing dimer stability or facilitating phosphorylation-independent activation. Understanding how these mutations alter SH2 domain dynamics and dimer interface stability provides insights for developing targeted inhibitors that specifically counteract pathogenic signaling.
Figure 2: SH2 Domain Therapeutic Targeting Approaches. Multiple strategies exploit different aspects of SH2 domain structure and function.
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Research Reagents for SH2 Domain Studies
| Reagent/Category | Specific Examples | Primary Application | Key Function in Research |
|---|---|---|---|
| Recombinant SH2 Domains | Wild-type and mutant SH2 domains (Btk, Fyn, STAT5) | Biophysical and biochemical studies | Folding, stability, and binding assays |
| Phosphopeptide Libraries | Combinatorial pY peptide libraries | Specificity profiling | Mapping SH2 domain binding preferences |
| NMR Isotope Labeling | 15N, 13C-labeled SH2 domains | NMR dynamics studies | Backbone and sidechain dynamics analysis |
| MD Simulation Software | GROMACS, AMBER, CHARMM | Molecular dynamics | Conformational sampling, allosteric pathways |
| Binding Assay Systems | Fluorescence polarization, SPR, ITC | Affinity and kinetics | Quantitative binding measurements |
| Engineered Protein Binders | Repebodies, monobodies, nanobodies | Allosteric inhibition | Targeting specific conformational states |
| Disease-Associated Mutants | XLA (Btk), leukemic (STAT5) mutants | Pathophysiological mechanisms | Linking dynamics to disease phenotypes |
SH2 domains exemplify how protein flexibility and dynamics enable sophisticated regulation of cellular signaling pathways. The STAT-SH2 domain in particular demonstrates how specialized structural adaptations facilitate controlled dimerization through a complex interface involving reciprocal phosphotyrosine recognition, hydrophobic packing, and unique intramolecular interactions. The dynamic nature of SH2 domains, mediated through flexible loops and internal allosteric networks, allows integration of multiple regulatory inputs and coordination with neighboring domains.
Understanding SH2 domain dynamics opens new avenues for therapeutic intervention, particularly through allosteric targeting and interface disruption strategies. As research methodologies continue to advance, particularly in molecular simulations and dynamics measurements, our ability to correlate SH2 domain flexibility with biological function will further improve. This knowledge will accelerate the development of novel therapeutics that selectively modulate SH2 domain interactions in disease states, offering promising approaches for targeting challenging pathologies driven by aberrant tyrosine kinase signaling.
Research Tools and Techniques: Probing SH2 Domain Dimerization Mechanisms
The Signal Transducer and Activator of Transcription (STAT) family of proteins represents a critical signaling node in cellular communication, directly influencing gene expression in response to cytokine and growth factor stimulation. Among the most critical steps in STAT protein activation is dimerization, which is essential for nuclear translocation and DNA binding. This process is mediated by the Src Homology 2 (SH2) domain, which recognizes and binds to phosphorylated tyrosine residues on a reciprocal STAT monomer. The structural basis for this interaction was revealed in the crystal structure of a tyrosine-phosphorylated STAT-1 dimer bound to DNA, which demonstrates how the dimer forms a C-shaped clamp around DNA, stabilized by specific interactions between the SH2 domain of one monomer and the phosphotyrosine-containing C-terminal segment of the other [32].
Understanding STAT dimerization is not merely of academic interest but has direct clinical implications. Dysregulation of JAK/STAT signaling is associated with various cancers and autoimmune diseases, making the protein-protein interactions mediated by the SH2 domain a promising therapeutic target [33]. This technical guide provides an in-depth examination of three principal methodologies used to characterize STAT dimerization: fluorescence co-localization, Förster Resonance Energy Transfer (FRET), and Analytical Ultracentrifugation (AUC). By framing these techniques within the context of STAT-SH2 domain research, this review serves as an essential resource for researchers and drug development professionals aiming to quantify and interrogate these critical interactions.
The STAT-SH2 Domain: Structure and Function
The SH2 domain is a modular protein interaction domain that arose within metazoan signaling pathways approximately 600 million years ago. In STAT proteins, it plays two non-redundant critical roles: it mediates the initial recruitment of STATs to activated cytokine receptors and facilitates the subsequent reciprocal dimerization of phosphorylated STAT proteins [5].
Structurally, STAT-type SH2 domains possess a central anti-parallel β-sheet (βB-βD strands) flanked by two α-helices (αA and αB), forming a characteristic αβββα motif. This structure creates two key binding pockets [5]:
- The pY (phosphate-binding) pocket: Formed by the αA helix, BC loop, and one face of the central β-sheet, this pocket engages the phosphotyrosine residue.
- The pY+3 (specificity) pocket: Created by the opposite face of the β-sheet, the αB helix, and adjacent loops, this pocket accommodates residues C-terminal to the phosphotyrosine, conferring binding specificity.
A unique feature of STAT-type SH2 domains is the presence of an additional α-helix (αB') in the C-terminal region of the pY+3 pocket, known as the evolutionary active region (EAR), which distinguishes them from Src-type SH2 domains that harbor a β-sheet in this region [5]. The integrity of the SH2 domain is maintained by a hydrophobic system of non-polar residues at the base of the pY+3 pocket, which stabilizes the β-sheet structure. The critical nature of this domain is highlighted by patient sequencing data, which identifies the SH2 domain as a hotspot for mutations in STAT3 and STAT5B, leading to either hyperactivated or refractory STAT mutants associated with various pathologies [5].
The following diagram illustrates the key structural features of the STAT SH2 domain and its role in dimerization:
Figure 1: STAT Dimerization Mechanism. The SH2 domain of one STAT monomer recognizes and binds to the phosphorylated tyrosine motif of a reciprocal STAT monomer, forming an active dimer capable of nuclear translocation.
Methodological Approaches for Assessing Dimerization
Fluorescence Co-localization
Principle and Application to STAT Dimerization
Fluorescence co-localization is a microscopy-based technique used to determine if two fluorescently labeled molecules occupy the same subcellular structures. While it lacks the resolution to directly prove molecular interaction, it can indicate association with common cellular compartments. In the context of STAT dimerization, co-localization can demonstrate that two differentially labeled STAT proteins converge in the cytoplasm upon pathway activation or co-accumulate in the nucleus following dimerization [34].
The fundamental approach involves labeling STAT proteins with distinct fluorophores (e.g., GFP and RFP) and visualizing their distribution in fixed or live cells via fluorescence microscopy. Co-localization is subjectively identified in merged images as structures whose color represents the combined contribution of both probes (e.g., yellow from red and green). However, this visual assessment is qualitative and can be ambiguous, as the combined color is highly dependent on the relative intensity and concentration of the two probes [34].
Quantitative Analysis and Key Considerations
For robust quantification, co-localization analysis must move beyond simple image merging. The recommended workflow involves:
- Image Acquisition: Collecting images with signals sufficient to distinguish from noise and background, and free of bleed-through between channels [34].
- Scatterplot Analysis: Plotting the intensity of each pixel in the red channel against its intensity in the green channel. Proportional co-distribution produces a scatterplot where points cluster around a straight line, whereas non-colocalizing probes form two separate clusters [34].
- Pearson's Correlation Coefficient (PCC): A statistical measure that quantifies the degree of linear correlation between pixel intensities in two channels. The formula is: ( PCC = \frac{\sum (Ri - \langle R \rangle)(Gi - \langle G \rangle)}{\sqrt{\sum (Ri - \langle R \rangle)^2 \sum (Gi - \langle G \rangle)^2}} ) where ( Ri ) and ( Gi ) are the intensity values of the red and green channels, respectively, and ( \langle R \rangle ) and ( \langle G \rangle ) are their mean values. A PCC value close to +1 indicates strong positive correlation, while values near 0 or negative indicate no or inverse correlation [34].
It is critical to distinguish between co-occurrence (simple spatial overlap) and correlation (proportional co-distribution). Two STAT probes might co-occur in the nucleus without their fluorescence intensities being correlated, which would suggest they are in the same compartment but not necessarily in a direct, stoichiometric complex [34].
Förster Resonance Energy Transfer (FRET)
Principle and Biosensor Design for STATs
FRET is a distance-dependent physical process where energy is non-radiatively transferred from an excited donor fluorophore to a nearby acceptor fluorophore. As FRET efficiency is inversely proportional to the sixth power of the distance between fluorophores, it is an exceptionally sensitive molecular ruler for measuring distances between 30 Å and 120 Å, ideally suited for studying dimerization [35].
FRET-based biosensors for STAT activity typically consist of a single polypeptide chain incorporating a donor-acceptor FRET pair (e.g., CFP and YFP) flanking a domain that undergoes a conformational change. For direct dimerization assays, a popular design is a unimolecular biosensor where ligand binding or phosphorylation induces an intramolecular conformational change that alters the distance/orientation between the FPs. The change in FRET efficiency serves as a quantitative readout of the molecular event [36].
A 2023 international blind study demonstrated that smFRET can achieve an inter-dye distance precision of ≤2 Å and an accuracy of ≤5 Å, confirming its reliability for characterizing structural dynamics in proteins [35].
Data Acquisition, Calibration, and Experimental Workflow
The most common method for measuring FRET is sensitized emission, which involves exciting the donor and measuring emission in both the donor and acceptor channels. The acceptor-to-donor signal ratio, or FRET ratio, is often used as a convenient surrogate for FRET efficiency. However, this ratio is influenced by laser intensity, detector sensitivity, and spectral crosstalk, complicating cross-experiment comparisons [36].
A robust calibration strategy involves using engineered "FRET-ON" (high FRET) and "FRET-OFF" (low FRET) standards, which can be imaged alongside the biosensor of interest. Theoretical and experimental work has shown that calibrating against both high and low FRET standards normalizes the FRET ratio, making it independent of imaging conditions and allowing for accurate determination of actual FRET efficiency [36]. The following workflow outlines the key steps in a calibrated FRET experiment:
Figure 2: Calibrated FRET Experimental Workflow. Incorporating calibration standards allows for robust, quantitative FRET measurements across different imaging sessions.
Critical correction factors must be determined using control samples (donor-only and acceptor-only) to account for i) spectral crosstalk (α), ii) differences in excitation fluxes and detection efficiencies (γ), and iii) direct acceptor excitation (δ) [35]. For single-molecule FRET (smFRET), the use of Alternating Laser Excitation (ALEX) or Pulsed Interleaved Excitation (PIE) is crucial for identifying and analyzing only molecules containing both active donor and acceptor fluorophores [35].
Analytical Ultracentrifugation (AUC)
Principle and Modes of Operation
Analytical Ultracentrifugation is a biophysical technique that leverages centrifugal force to characterize macromolecular properties in free solution without a solid matrix. It provides unparalleled information about hydrodynamic and thermodynamic properties, making it ideal for studying protein self-association, such as STAT dimerization [37]. Two primary modes are used:
- Sedimentation Velocity (SV): In this hydrodynamic method, high centrifugal force causes molecules to migrate based on their mass, density, and shape. The temporal evolution of the concentration profile is monitored, allowing for the determination of sedimentation coefficients and the identification of different species in a mixture. For static associations (slowly reversible), distinct boundaries for monomer and oligomer are observed, whereas dynamic associations (rapidly reversible) manifest as a single, broad boundary [38] [37].
- Sedimentation Equilibrium (SE): In this thermodynamic method, a lower centrifugal force is applied until equilibrium is reached where sedimentation is balanced by diffusion. The resulting concentration profile depends on the buoyant molar mass of the solute(s). SE is exquisitely sensitive to changes in molecular weight, allowing for the determination of association constants and stoichiometries for interacting systems [37] [38].
AUC has been successfully applied to characterize STAT proteins, providing a quantitative understanding of dimer stability and conformational transitions associated with activation [39].
Experimental Design and Data Analysis
Modern SV data analysis often involves numerical solutions of the Lamm equation, which describes the evolution of concentration profiles in the centrifugal field. Fitting experimental data to these solutions allows for the determination of sedimentation coefficient distributions, ( c(s) ), which deconvolute the diffusion, revealing the number and proportion of sedimenting species [37] [38].
For multi-component interactions, multi-signal SV is a powerful advancement. This technique utilizes differences in the spectral properties of individual components (e.g., STAT1 and STAT3). By globally analyzing data from multiple optical detection systems (e.g., UV-Vis absorbance and interference optics), it is possible to determine the stoichiometry and affinity of hetero-complex formation [37].
For SE experiments in dilute solutions, the data for a self-associating system like a STAT monomer-dimer equilibrium can be described by: ( c(r) = c{m}(r0) \exp[M{1}(1-\overline{v}\rho)\omega^2 (r^2 - r0^2)/(2RT)] + K{12} c{m}(r0)^2 \exp[2M{1}(1-\overline{v}\rho)\omega^2 (r^2 - r0^2)/(2RT)] ) where ( c(r) ) is the total concentration at radius ( r ), ( cm ) is the monomer concentration, ( M1 ) is the monomer molecular weight, ( \overline{v} ) is the partial specific volume, ( \rho ) is the solvent density, ( \omega ) is the angular velocity, ( R ) is the gas constant, ( T ) is the absolute temperature, and ( K{12} ) is the equilibrium association constant [37].
Comparative Analysis of Techniques
Table 1: Comparison of Techniques for Studying STAT Dimerization
| Feature | Fluorescence Co-localization | FRET | Analytical Ultracentrifugation |
|---|---|---|---|
| Primary Information | Spatial overlap in cellular compartments | Distance (30-120 Å), conformational dynamics, interaction proximity | Hydrodynamic properties, molar mass, stoichiometry, equilibrium constants |
| Resolution | ~200 nm (diffraction-limited) | Molecular scale (Ångströms) | Molecular scale (Sedimentation coefficient, S) |
| Cellular Context | Yes (in situ/in vivo) | Yes (in situ/in vivo with biosensors) | No (in vitro, purified components) |
| Throughput | Moderate to High | Moderate | Low |
| Sample Requirement | Fluorescently tagged proteins | Fluorescently tagged proteins/biosensors | Purified protein, no labeling required |
| Key Quantitative Outputs | Pearson's Correlation Coefficient, Scatterplots | FRET Efficiency, Distances, Dynamics | Sedimentation coefficient (s), Association constant (K~d~), Molecular weight |
| Application to STAT SH2 | Visualize co-transport to nucleus; infer interaction | Directly probe SH2-pY interaction and conformational changes | Quantify dimer stability and affinity in solution |
Research Reagent Solutions
Table 2: Essential Research Reagents for STAT Dimerization Studies
| Reagent / Tool | Function / Description | Example Application |
|---|---|---|
| STAT SH2 Domain Constructs | Recombinant proteins containing the isolated SH2 domain. | Used in FP/AUC assays to study binding thermodynamics without full-length protein complexity [40]. |
| Phosphopeptide Ligands | Fluorophore-labeled or unlabeled peptides containing the phosphotyrosine motif (e.g., GpYLPQNID). | Probes for binding assays (FP, SPR) to measure SH2 domain affinity and for competitive inhibition studies [40]. |
| FRET Biosensors | Genetically encoded constructs (e.g., CFP-YFP flanking a STAT conformational sensor). | Live-cell imaging of STAT activation and dimerization dynamics [36]. |
| Fluorescence Polarization Tracers | Small, fluorophore-labeled phosphopeptides. | High-throughput screening for SH2 domain inhibitors in 384-well format [40]. |
| Calibration Standards (FRET-ON/OFF) | Engineered constructs with fixed high or low FRET efficiency. | Normalization of FRET ratios across different imaging sessions and instruments [36]. |
| Double-Cysteine STAT Mutants | STAT variants with two engineered cysteine residues for site-specific dye labeling. | For smFRET studies to ensure specific fluorophore attachment at desired positions [35]. |
The rigorous characterization of STAT dimerization, driven by SH2 domain interactions, is foundational to understanding cellular signaling and developing targeted therapeutics. Each technique discussed—co-localization, FRET, and AUC—provides a unique and complementary lens through which to study this process.
Fluorescence co-localization offers the invaluable ability to place protein interactions within the morphological context of the cell, suggesting functional consequences in specific compartments. FRET provides unparalleled sensitivity for probing molecular-scale distances and real-time conformational dynamics in living cells, especially when paired with robust calibration standards. Analytical Ultracentrifugation remains the gold standard for in vitro quantitative characterization, yielding precise thermodynamic and hydrodynamic parameters of interactions without the need for labeling.
The future of STAT dimerization research lies in an integrative approach. Combining these methodologies, along with structural techniques like X-ray crystallography [32] and emerging technologies, will provide a holistic view of STAT protein function. This multi-faceted strategy will continue to drive discoveries in basic science and accelerate the development of novel therapeutics targeting the JAK/STAT pathway in cancer and autoimmune diseases.
The signal transducer and activator of transcription (STAT) family of proteins are fundamental cytoplasmic transcription factors that transduce signals from cytokine and growth factor receptors to the nucleus, regulating genes involved in proliferation, survival, and immune responses [4] [41]. The canonical activation of STATs, particularly STAT3 and STAT5, relies on a critical protein-protein interaction mediated by their Src Homology 2 (SH2) domains [5]. The SH2 domain is an approximately 100-amino-acid modular unit that specifically recognizes and binds to phosphorylated tyrosine (pY) motifs [8] [16]. In STAT biology, this domain has a dual function: it initially recruits monomeric STATs to activated cytokine receptors via the receptor's phosphorylated tyrosine residues, and subsequently facilitates STAT dimerization through a reciprocal phosphotyrosine-SH2 domain interaction between two STAT monomers [42] [5].
Upon phosphorylation at a conserved C-terminal tyrosine residue (e.g., Tyr705 in STAT3), two STAT monomers form a parallel dimer through a specific molecular interaction where the SH2 domain of one monomer binds the phosphotyrosine motif of the other, and vice versa [32] [5]. This dimerization event is essential for the nuclear translocation of STATs and their binding to specific DNA response elements to regulate transcription [42] [41]. The centrality of the SH2 domain in this process makes it a hotspot for disease-associated mutations and a prime target for therapeutic intervention in cancers and immune disorders [5]. This technical guide details the application of X-ray crystallography and Molecular Dynamics (MD) simulations, two pivotal structural biology techniques, for investigating the mechanism of STAT dimerization and for informing targeted drug discovery.
Fundamental Principles of the STAT-type SH2 Domain
Structural Architecture and Classification
SH2 domains are a metazoan-specific adaptation that share a conserved core fold despite significant sequence divergence. All SH2 domains assume a characteristic "sandwich" structure consisting of a central three-stranded antiparallel beta-sheet (βB-βC-βD) flanked by two alpha helices (αA and αB) [8] [5]. This creates an αβββα motif that forms two key binding pockets: a phosphate-binding (pY) pocket that engages the phosphotyrosine moiety, and a specificity (pY+3) pocket that recognizes residues C-terminal to the pY, conferring selectivity for distinct peptide motifs [5] [16].
STAT-type SH2 domains are a distinct subgroup characterized by unique structural features that differentiate them from Src-type SH2 domains. Most notably, STAT-type domains lack the βE and βF strands found in Src-type domains and instead possess an additional α-helix (αB') in the C-terminal region, known as the evolutionary active region (EAR) [8] [5]. This structural adaptation is critical for its primary function in mediating STAT dimerization. The pY pocket contains a highly conserved arginine residue (from the FLVR motif) that forms a salt bridge with the phosphate group of the phosphotyrosine, a interaction fundamental to both canonical and STAT-type SH2 domains [8] [16].
The SH2 Domain in STAT Dimerization and Activation
The STAT activation cascade is a multi-step process visually summarized below. The SH2 domain is the linchpin in the dimerization step, which is the focus of structural biology investigations.
X-ray Crystallography for Elucidating STAT-SH2 Domain Architecture
X-ray crystallography is a powerful technique for determining the three-dimensional atomic structure of macromolecules. It involves purifying the protein of interest, growing a highly ordered crystal, collecting diffraction data from the crystal exposed to an X-ray beam, and using computational methods to generate an electron density map to build and refine an atomic model [32]. For STAT SH2 domains, this technique has been instrumental in revealing the precise molecular interactions that govern dimerization.
A landmark study used X-ray crystallography to solve the structure of a tyrosine-phosphorylated STAT-1 homodimer bound to DNA at 2.9 Å resolution [32]. This structure revealed that the STAT-1 dimer forms a C-shaped clamp around DNA, which is stabilized by reciprocal and highly specific interactions between the SH2 domain of one monomer and the phosphorylated C-terminal tyrosine segment of the other. The structure clearly showed that the phosphotyrosine-binding site of the SH2 domain in each monomer is coupled structurally to the DNA-binding domain, suggesting a role for the SH2-pY interaction in stabilizing DNA-binding elements [32].
Experimental Protocol for STAT SH2 Domain Crystallography
A typical workflow for determining a STAT SH2 domain structure involves the following key steps [32] [5]:
- Protein Expression and Purification: The STAT SH2 domain (or full-length protein) is cloned, expressed in a system like insect or mammalian cells, and purified using affinity chromatography (e.g., Ni-NTA for his-tagged proteins) and size-exclusion chromatography.
- Crystallization: The purified protein is concentrated and subjected to crystallization trials using robotic screens. Crystals of the STAT SH2 domain are often grown in the presence of a bound phosphopeptide to stabilize the active conformation.
- Data Collection and Processing: A single crystal is flash-cooled in liquid nitrogen. X-ray diffraction data are collected at a synchrotron facility. The diffraction patterns are processed and integrated to obtain structure factor amplitudes.
- Phase Determination and Model Building: Phases are determined by molecular replacement using a known SH2 domain structure as a search model. An initial atomic model is built into the experimental electron density map.
- Refinement and Validation: The model is iteratively refined against the diffraction data to improve its fit and validated using geometric and stereochemical criteria.
Table 1: Key Insights from X-ray Crystallography of STAT SH2 Domains
| Structural Insight | Description | Experimental Basis |
|---|---|---|
| Dimerization Interface | Reciprocal SH2-pY interaction between two STAT monomers. | Crystal structure of phosphorylated STAT-1 dimer [32]. |
| DNA Binding Clamp | Dimer forms a contiguous C-shaped clamp around DNA. | STAT-1 dimer-DNA co-crystal structure [32]. |
| STAT-type Specificity | Presence of C-terminal αB' helix and absence of βE/F strands. | Comparative structural analysis of SH2 domains [8] [5]. |
| Ligand Binding Pockets | Definition of the pY (phosphate-binding) and pY+3 (specificity) pockets. | Structures of SH2 domains bound to phosphopeptides [5] [16]. |
Molecular Dynamics Simulations for Probing STAT-SH2 Dynamics
While X-ray crystallography provides a static, high-resolution snapshot, Molecular Dynamics (MD) simulations model the dynamic behavior of atoms and molecules over time, typically on a picosecond-to-microsecond scale. MD simulations use Newton's equations of motion to calculate the trajectories of every atom in a system, which consists of the protein solvated in water molecules and ions [43]. This technique is exceptionally valuable for capturing the flexibility, conformational changes, and energetic landscapes of the STAT SH2 domain, which are often crucial for function but obscured in crystal structures.
For instance, MD simulations have revealed that STAT SH2 domains exhibit significant flexibility, even on sub-microsecond timescales. A key finding is that the accessible volume of the critical pY binding pocket can vary dramatically, a feature that is often not captured in a single crystal structure [5]. This dynamic behavior has profound implications for drug discovery, as it reveals that the binding site is not a static cavity but a malleable one.
Experimental Protocol for MD Simulations of the STAT SH2 Domain
A standard MD simulation protocol for studying the STAT SH2 domain involves [43] [5]:
- System Preparation: An initial structure (e.g., from an X-ray crystal structure or a homology model) is placed in a simulation box filled with explicit water molecules (e.g., TIP3P model). Ions are added to neutralize the system's charge and mimic physiological salt concentration.
- Energy Minimization: The system's energy is minimized to remove any steric clashes introduced during the setup, ensuring stable initial conditions for the simulation.
- Equilibration: The system is gradually heated to the target temperature (e.g., 310 K) and the pressure is equilibrated to 1 bar. Positional restraints on the protein heavy atoms are typically applied and then gradually released.
- Production Run: An unrestrained simulation is run for a defined period (nanoseconds to microseconds), during which atomic coordinates and velocities are saved at regular intervals for subsequent analysis.
- Trajectory Analysis: The saved trajectories are analyzed to compute properties such as root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), radius of gyration, hydrogen bonding patterns, and free-energy landscapes.
A comparative study on a different protein system, acid-beta-glucosidase, demonstrated the complementary nature of MD and crystallography. While the X-ray study identified a conformational change in an active site loop, the MD simulation provided unique additional data on the flexibility of another loop and the catalytic residues, which were not observable in the static crystal structure [43]. This underscores MD's unique capability to probe protein dynamics and flexibility.
Integrated Structural Approach in STAT-Targeted Drug Discovery
Synergistic Application of Techniques
The most powerful insights are gained when X-ray crystallography and MD simulations are used in an integrated fashion. Crystallography provides the high-resolution structural framework and starting coordinates for simulations, while MD validates the stability of the observed conformations, identifies allosteric pockets, and models the molecular basis for the functional impact of mutations [43] [5]. This synergy is critical for targeting the STAT3 SH2 domain in cancer therapy, as illustrated below.
Case Study: Discovery of STAT3 SH2 Domain Inhibitors
The discovery of novel STAT3 inhibitors, such as the delavatine A stereoisomers (compounds 323-1 and 323-2), exemplifies this integrated approach [42]:
- Computational Docking: Initial in silico docking of compounds into the STAT3 SH2 domain structure predicted binding to three key subpockets.
- MD Simulations: Simulations were likely used to refine the binding poses and assess the stability of inhibitor-domain complexes.
- Biophysical Validation: The direct interaction and competitive inhibition were confirmed experimentally using Fluorescence Polarization (FP) assays, which showed the compounds abrogated the interaction between STAT3 and a SH2-binding peptide (GpYLPQTV).
- Functional Validation: Drug Affinity Responsive Target Stability (DARTS) and co-immunoprecipitation assays confirmed the compounds directly target the SH2 domain and disrupt STAT3 dimerization more potently than a reference inhibitor (S3I-201) [42].
Table 2: Key Reagent Solutions for STAT SH2 Domain Dimerization Research
| Research Reagent / Assay | Function in STAT-SH2 Research |
|---|---|
| Recombinant STAT SH2 Domain Protein | Purified protein for in vitro binding assays, crystallization trials, and biophysical characterization. |
| Phosphorylated Peptide Ligands (e.g., GpYLPQTV) | Mimic the native phosphotyrosine ligand to study binding affinity and specificity in FP and SPR assays. |
| Fluorescence Polarization (FP) Assay | Quantifies the binding affinity and competitive inhibition of small molecules by measuring changes in fluorescence polarization. |
| Drug Affinity Responsive Target Stability (DARTS) | Identifies direct protein-small molecule interactions based on protease resistance conferred by ligand binding. |
| Co-immunoprecipitation (Co-IP) | Assesses the disruption of STAT dimerization in a cellular context. |
| STAT3 Reporter Luciferase Assay | Measures the functional consequence of SH2 domain inhibition on STAT3-mediated transcriptional activity. |
| Crystal Screen Kits (e.g., from Hampton Research) | Commercial sparse-matrix screens for initial crystallization condition identification. |
Analyzing Disease-Associated Mutations
The STAT SH2 domain is a mutational hotspot in diseases like autosomal-dominant hyper IgE syndrome (AD-HIES) and various leukemias [5]. Integrated structural biology is essential for understanding these mutations. X-ray structures can show the spatial location of a mutated residue (e.g., S614R in STAT3), while MD simulations can reveal how the mutation alters local flexibility, hydrogen-bonding networks, or allosterically affects the pY pocket's conformation, leading to either gain-of-function or loss-of-function phenotypes [43] [5]. This mechanistic understanding is a critical first step towards developing targeted interventions.
The Src Homology 2 (SH2) domain is a critical modular unit within metazoan signaling pathways, serving as a specialized phosphotyrosine-binding module that directs protein-protein interactions in response to tyrosine phosphorylation [14]. In Signal Transducer and Activator of Transcription (STAT) proteins, the SH2 domain plays an indispensable role in molecular activation, facilitating JAK-mediated phosphorylation, STAT dimerization, and nuclear translocation of phosphorylated STAT dimers to drive transcription of target genes [5] [32]. The structural integrity of the SH2 domain is therefore essential for normal STAT function, and its disruption has been directly linked to numerous human diseases.
Recent advances in genomic sequencing have revealed the SH2 domain as a mutational hotspot in the STAT protein landscape [5]. Patient sequencing data has identified numerous point mutations within the STAT3 and STAT5 SH2 domains that result in either hyperactivated or refractory STAT mutants, contributing to various pathologies including immunodeficiencies, leukemias, and solid tumors [5] [44]. Understanding the molecular and biophysical impact of these disease-associated mutations reveals convergent mechanisms of action that can facilitate targeted therapeutic interventions [5].
This technical guide provides an in-depth analysis of the STAT-SH2 domain mutational landscape, with a specific focus on how these mutations disrupt normal STAT dimerization and function. We present comprehensive structural data, detailed experimental methodologies, and visualization tools to support research efforts aimed at deciphering SH2 domain pathology and developing targeted therapeutic strategies.
STAT-SH2 Domain Structure and Function
Structural Architecture of STAT-Type SH2 Domains
SH2 domains are approximately 100 amino acids in length and share a conserved structural fold that consists of a central anti-parallel β-sheet flanked by two α-helices, forming an αβββα motif [5] [14]. The central β-sheet partitions the domain into two functionally distinct subpockets:
- pY pocket (phosphate-binding pocket): Formed by the αA helix, BC loop, and one face of the central β-sheet, this pocket contains highly conserved residues that interact directly with the phosphotyrosine moiety [5].
- pY+3 pocket (specificity pocket): Created by the opposite face of the β-sheet along with residues from the αB helix and CD and BC* loops, this pocket engages residues C-terminal to the phosphotyrosine to confer binding specificity [5].
STAT-type SH2 domains possess distinctive features that differentiate them from Src-type SH2 domains. Most notably, STAT-type domains contain an α-helix (αB') at their C-terminus, as opposed to the β-sheet (βE and βF) found in Src-type domains [5] [24]. This structural variation, located in what is termed the evolutionary active region (EAR), contributes to the unique functional properties of STAT SH2 domains [5].
Table 1: Key Structural Motifs in STAT SH2 Domains
| Structural Element | Location | Functional Role | Conservation |
|---|---|---|---|
| αA helix | N-terminal | Forms part of pY pocket | High |
| βB strand | Central | Contains conserved arginine for pTyr binding | Very high |
| BC loop | Between βB-βC | Contributes to pY pocket formation | Moderate |
| αB helix | C-terminal | Part of pY+3 pocket and dimerization interface | Moderate |
| αB' helix | C-terminal | STAT-type specific feature; involved in dimerization | STAT-specific |
| BC* loop | Between αB-αC | Participates in SH2-mediated STAT dimerization | Variable |
Role in STAT Activation and Dimerization
The SH2 domain is fundamental to the canonical STAT activation pathway. In response to cytokine or growth factor stimulation, SH2 domains mediate recruitment of STAT proteins to phosphorylated tyrosine motifs on activated receptors [5]. This positioning allows JAK kinases to phosphorylate a conserved tyrosine residue in the STAT C-terminal tail.
Following phosphorylation, STAT proteins form reciprocal dimers through phosphotyrosine-SH2 domain interactions, where the phosphorylated tyrosine of one STAT molecule binds to the SH2 domain of its partner, and vice versa [32] [30]. The crystal structure of tyrosine-phosphorylated STAT-1 dimer bound to DNA reveals that the dimer forms a contiguous C-shaped clamp around DNA, stabilized by specific interactions between the SH2 domain of one monomer and the phosphorylated C-terminal segment of the other [32].
This dimerization is essential for nuclear translocation and DNA binding, enabling STATs to function as transcription factors regulating genes involved in proliferation, survival, and differentiation [5]. Recent research on STAT5 has identified additional intramolecular hydrophobic interactions involving residues adjacent to the phosphotyrosine motif that are critical for stable dimer formation [30].
Figure 1: STAT Protein Activation Pathway. Cytokine binding triggers receptor activation and JAK-mediated phosphorylation. STAT proteins are recruited via SH2 domains, become phosphorylated, and form reciprocal dimers through SH2-phosphotyrosine interactions before nuclear translocation.
Disease-Associated Mutations in STAT SH2 Domains
STAT3 SH2 Domain Mutations
The STAT3 SH2 domain represents a significant mutational hotspot in various human diseases. Sequencing analyses of patient samples have identified numerous point mutations that cluster in specific regions of the SH2 domain, with distinct pathological consequences depending on the nature and location of the mutation [5].
Table 2: Disease-Associated Mutations in STAT3 SH2 Domain
| Mutation | Position | Location | Pathology | Type | Functional Effect |
|---|---|---|---|---|---|
| K591E/M | αA2 | pY pocket | AD-HIES | Germline | Loss-of-function |
| R609G | βB5 | pY pocket | AD-HIES | Germline | Loss-of-function |
| S611N/G/I | βB7 | pY pocket | AD-HIES | Germline | Loss-of-function |
| S614R | BC3 | pY pocket | T-LGLL, NK-LGLL, ALK-ALCL, HSTL | Somatic | Gain-of-function |
| E616G/K | BC5 | pY pocket | DLBCL, NKTL | Somatic | Gain-of-function |
| G617E/V/R | BC6 | pY pocket | AD-HIES | Germline | Loss-of-function |
Loss-of-function mutations in STAT3 are predominantly associated with autosomal-dominant hyper IgE syndrome (AD-HIES), an immunological disorder characterized by recurrent staphylococcal infections, eczema, and eosinophilia [5]. These mutations typically disrupt conserved residues in the pY pocket, impairing phosphotyrosine binding and STAT3 dimerization. The resulting defect in STAT3-mediated Th17 T-cell response reduces immunological competence [5].
Conversely, gain-of-function mutations such as S614R and E616K are found in hematologic malignancies including T-cell large granular lymphocytic leukemia (T-LGLL), natural killer T-cell lymphoma (NKTL), and other lymphoid cancers [5]. These mutations often enhance STAT3 dimerization and transcriptional activity, promoting cell proliferation and survival.
STAT5B SH2 Domain Mutations
STAT5B SH2 domain mutations demonstrate how different substitutions at the same residue can produce opposing functional effects. The most extensively characterized mutations occur at tyrosine 665 (Y665), which can be substituted with either phenylalanine (Y665F) or histidine (Y665H) [45] [44].
The Y665F mutation is a well-established gain-of-function variant identified in T-cell leukemias, including T-LGLL and T-PLL [44]. This mutation enhances STAT5 phosphorylation, DNA binding, and transcriptional activity after cytokine activation, leading to accumulation of CD8+ effector and memory T cells and altered CD8+/CD4+ T-cell ratios [44].
In contrast, the Y665H mutation functions as a loss-of-function variant, diminishing CD8+ effector and memory T cells and CD4+ regulatory T cells [44]. In mammary gland development, STAT5B Y665H impairs alveolar differentiation and lactation, while Y665F accelerates mammary development during pregnancy [45].
Molecular dynamics simulations suggest that these opposing effects stem from how each substitution alters the hydrophobic interactions at the dimer interface. The phenylalanine substitution in Y665F enhances hydrophobic contacts, stabilizing the active dimer, while histidine introduces a polar residue that disrupts these critical interactions [30].
Experimental Approaches for Characterization
Structural Analysis Techniques
X-ray Crystallography has provided foundational insights into STAT SH2 domain structure and function. The crystal structure of tyrosine-phosphorylated STAT-1 dimer bound to DNA revealed the molecular details of reciprocal SH2-phosphotyrosine interactions that stabilize the active dimer [32]. This approach remains valuable for determining high-resolution structures of mutant SH2 domains.
Molecular Dynamics (MD) Simulations enable researchers to model the structural consequences of SH2 domain mutations. For STAT5, MD simulations of the dimer interface identified a novel intramolecular interaction mediated through F706, adjacent to the phosphotyrosine motif, and a unique hydrophobic interface on the SH2 domain surface [30]. These simulations revealed how mutations at Y665 alter the stability of the dimerization interface, helping explain their opposing functional effects.
Functional Characterization Methods
Deep Mutational Scanning represents a powerful high-throughput approach for comprehensively characterizing mutational effects. Recently applied to the multi-domain phosphatase SHP2 (which contains two SH2 domains), this method enables profiling of thousands of mutants in parallel, revealing mechanistically diverse mutational effects and identifying key intra- and inter-domain interactions [46]. Similar approaches could be applied systematically to STAT SH2 domains.
SH2 Domain Specificity Profiling using peptide microarrays has enabled large-scale characterization of SH2-phosphopeptide interactions. One study developed high-density peptide chip technology containing nearly the entire complement of human tyrosine phosphopeptides, probing the affinity of 70 different SH2 domains and identifying thousands of putative interactions [47] [48]. This resource helps contextualize how disease-associated mutations might alter SH2 domain specificity.
In Vivo Modeling
CRISPR/Cas9 and Base Editing in mice allows introduction of human disease-associated mutations into the mouse genome for physiological studies. The STAT5B Y665F and Y665H mutations have been modeled using these approaches, with ABE mRNA and sgRNA co-microinjected into fertilized eggs or electroporated into zygotes [45]. This enables investigation of mutation impacts in key target tissues under different physiological states.
Transcriptomic and Epigenomic Analyses of mutant mouse models reveal how SH2 domain mutations alter gene regulatory networks. RNA-seq and enhancer mapping (e.g., ATAC-seq) of STAT5B Y665F and Y665H mutants demonstrated distinct effects on enhancer establishment and transcriptional programs in T cells and mammary tissue [45] [44].
Figure 2: Experimental Approaches for SH2 Domain Mutation Analysis. Interdisciplinary methods spanning structural, functional, and in vivo analysis provide comprehensive characterization of SH2 domain mutations.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for SH2 Domain Studies
| Reagent/Tool | Specifications | Application | Key Features |
|---|---|---|---|
| SH2 Domain Profiling Chip | 6202 phosphopeptides, 13 residues long, pTyr in middle | Specificity profiling | High-density peptide array; quantitative binding data |
| GST-tagged SH2 Domains | 70 human SH2 domains as GST fusions | Binding assays | Soluble expression; tag for purification/detection |
| STAT SH2 Domain Mutant Libraries | Saturation mutagenesis coverage | Deep mutational scanning | Comprehensive variant analysis; structure-function maps |
| CRISPR Base Editors | ABE7.10 mRNA with sgRNA | In vivo modeling | Direct single nucleotide conversion without double-strand breaks |
| Phosphospecific STAT Antibodies | Anti-pY694/699 for STAT5, pY705 for STAT3 | Activation assessment | Pathway activation readout; flow cytometry, Western blot |
| SH2 Domain Inhibitors | Small molecules targeting pY/pY+3 pockets | Therapeutic targeting | Disrupt specific protein interactions |
Discussion and Future Directions
The mapping of disease-associated mutations in the STAT SH2 domain landscape reveals fundamental principles of protein structure-function relationships and their disruption in human disease. Several key patterns emerge from this analysis:
First, the genetic volatility of specific regions in the SH2 domain can result in either activating or deactivating mutations at the same site, underscoring the delicate evolutionary balance of wild-type STAT structural motifs in maintaining precise levels of cellular activity [5]. The opposing effects of Y665F and Y665H mutations in STAT5B exemplify how subtle changes in chemical properties at critical positions can dramatically alter function.
Second, mutations at structurally conserved positions often produce consistent pathological effects across STAT family members. For example, residues involved in phosphotyrosine coordination (e.g., R618 in STAT5B, corresponding to R609 in STAT3) typically cause loss-of-function when mutated, while mutations in surrounding regions can yield gain-of-function variants that promote cytokine-independent activation [5] [30].
Third, the flexibility and dynamics of STAT SH2 domains present both challenges and opportunities for therapeutic development. Molecular dynamics simulations reveal that STAT SH2 domains exhibit considerable flexibility even on sub-microsecond timescales, with dramatic variations in the accessible volume of the pY pocket [5]. This flexibility must be accounted for in drug discovery efforts.
Future research directions should include more comprehensive deep mutational scanning of STAT SH2 domains to systematically map mutational effects, enhanced structural studies of disease-associated variants, and continued development of small molecule inhibitors targeting pathological SH2 domain interactions. The integration of these approaches will advance both our understanding of STAT biology and our ability to target these transcription factors therapeutically in cancer and immune disorders.
The Signal Transducer and Activator of Transcription (STAT) family of proteins has traditionally been studied through the lens of its canonical activation pathway, where tyrosine phosphorylation by Janus kinases (JAKs) triggers STAT dimerization via reciprocal Src Homology 2 (SH2) domain-phosphotyrosine interactions, leading to nuclear translocation and transcriptional activation [49]. However, emerging research has established that unphosphorylated STATs (U-STATs) possess biologically significant functions independent of tyrosine phosphorylation, engaging in non-canonical dimerization and nuclear roles that expand their functional repertoire in both normal physiology and disease states [50] [4] [49]. This paradigm shift reveals that STAT proteins function not merely as inducible signaling molecules but as versatile regulators of gene expression through mechanisms distinct from their phosphorylated counterparts.
The STAT-SH2 domain, traditionally considered essential for phosphotyrosine-mediated dimerization in canonical signaling, also plays roles in the context of these non-canonical functions, though the dimerization mechanisms differ substantially [51] [15] [52]. This technical guide provides a comprehensive examination of U-STAT dimerization and nuclear functions, framing these findings within the broader context of STAT-SH2 domain research to inform future investigative and therapeutic approaches.
Structural Mechanisms of Unphosphorylated STAT Dimerization
Alternative Dimerization Interfaces
The structural biology of unphosphorylated STAT dimers reveals fundamental differences in dimerization interfaces compared to their phosphorylated counterparts. While phosphorylated STATs dimerize through reciprocal SH2 domain-phosphotyrosine interactions, unphosphorylated STATs utilize distinct protein domains to form dimers.
Table 1: Comparison of Phosphorylated and Unphosphorylated STAT Dimerization Mechanisms
| Feature | Phosphorylated STAT Dimers | Unphosphorylated STAT Dimers |
|---|---|---|
| Primary Dimerization Interface | Reciprocal SH2 domain-phosphotyrosine interactions [49] | β-barrel and four-helix bundle domains [53] |
| Dimerization Trigger | Tyrosine phosphorylation (typically around Y700) [4] | Constitutive, phosphorylation-independent [53] |
| SH2 Domain Role | Essential for dimer formation [15] | Not primary for dimer interface [53] |
| Stability | Transient, phosphorylation-dependent | Stable in cytoplasmic latency [53] |
| Nuclear Translocation Efficiency | High, active import [4] | Variable, distinct mechanisms [4] |
Crystallographic studies of unphosphorylated STAT5a homodimers lacking the N-terminal domain and C-terminal transactivation domain have revealed a completely different dimerization mode mediated through interactions between their β-barrel and four-helix bundle domains rather than SH2 domains [53]. This structural arrangement maintains STATs in a latent dimeric configuration in the cytoplasm before activation, challenging the previous paradigm of exclusively monomeric latent STATs.
Role of the SH2 Domain in Non-Canonical Contexts
While not primary for U-STAT dimerization, the SH2 domain maintains functional importance in non-canonical signaling contexts through alternative mechanisms:
- Phosphotyrosine-independent interactions: Certain SH2 domains can engage in weak but functionally significant interactions with unphosphorylated targets, potentially facilitating non-canonical protein complexes [15].
- Structural stabilization: The SH2 domain contributes to the overall structural integrity of U-STAT dimers even when not directly mediating dimerization [52].
- Receptor docking capability: U-STATs maintain the capacity for receptor interaction via SH2 domains, potentially enabling phosphorylation-independent signaling functions [51].
The evolutionary conservation of SH2 domains across STAT family members underscores their fundamental importance, with over 100 human proteins containing these domains that typically recognize phosphotyrosine-containing motifs [15]. This conservation suggests constrained evolutionary flexibility despite the emergence of phosphorylation-independent functions.
Figure 1: Comparative Pathways of Canonical and Non-Canonical STAT Dimerization. Canonical signaling requires tyrosine phosphorylation and SH2 domain-mediated dimerization, while non-canonical pathways utilize alternative structural domains for dimerization and distinct nuclear import mechanisms.
Nuclear Functions of Unphosphorylated STATs
Mechanisms of Nuclear Translocation
Unphosphorylated STATs employ distinct nuclear transport mechanisms compared to their phosphorylated counterparts. Unlike phosphorylated STAT dimers that utilize active import via importin proteins, U-STATs shuttle between cytoplasm and nucleus through alternative pathways:
- Importin-independent transport: U-STAT1 undergoes nuclear translocation through direct interaction with nucleoporins rather than traditional importin receptors [49].
- Selective importin usage: U-STAT3 nuclear import is mediated specifically by importin-α3 and importin-α6, with importin-α3 serving as the primary transporter due to its ubiquitous expression [4].
- Energy-independent shuttling: The nuclear-cytoplasmic transport of U-STATs occurs independently of metabolic energy in contrast to the active transport of phosphorylated STAT dimers [4].
The coiled-coil domain of STAT3 contains critical nuclear localization signals (NLS) between amino acids 150-162 that are recognized by specific import carriers [4]. This domain-specific recognition mechanism explains the differential nuclear transport properties among STAT family members and their phosphorylation states.
Gene Regulatory Mechanisms
Once in the nucleus, U-STATs regulate gene expression through diverse molecular mechanisms that differ fundamentally from canonical STAT functions:
Table 2: Nuclear Functions of Unphosphorylated STAT3
| Function | Mechanism | Biological Outcome |
|---|---|---|
| DNA Binding | Binds GAS elements as dimers and monomers; recognizes AT-rich sequences and specific DNA structures [4] | Transcriptional activation of distinct gene sets |
| Chromatin Organization | Binds DNA nodes and 4-way junctions; recognizes single-stranded spacers within cruciform structures [4] | Genome architecture modulation |
| Disulfide Bridge Regulation | Cys367-Cys542 disulfide bridge stabilizes dimeric form essential for DNA-binding [4] | Enhanced DNA binding capacity |
| Heterochromatin Influence | Modulates heterochromatin stability; affects epigenetic landscape [54] | Altered accessibility of transcription factors |
U-STAT3 accumulates in response to strong STAT3 gene activation by phosphorylated STAT3 dimers, creating a self-regulatory feedback loop where canonical signaling induces non-canonical effectors [4]. This accumulation drives gene expression through novel mechanisms that extend beyond traditional transcriptional activation, including chromatin organization and genome architecture modulation.
U-STATs can bind both similar and distinct DNA sequences compared to their phosphorylated counterparts. For example, U-STAT3 preferentially binds AT-rich DNA sequences and specific DNA structures, leading to heterochromatin formation and gene silencing [49]. This functional diversity enables a single STAT protein to regulate broad transcriptional programs through phosphorylation-dependent and independent mechanisms.
Experimental Approaches for Studying Non-Canonical STAT Functions
Methodologies for Investigating U-STAT Dimerization
Structural Analysis Techniques
X-ray Crystallography:
- Protein Preparation: Express and purify recombinant STAT fragments lacking N-terminal and C-terminal domains (e.g., STAT5a ΔNTD ΔTAD) using baculovirus or bacterial expression systems [53].
- Crystallization: Utilize vapor diffusion methods with screening solutions containing PEGs of various molecular weights to obtain crystals diffracting to 3.0-3.5 Å resolution.
- Data Collection and Structure Determination: Collect diffraction data at synchrotron sources and solve structures using molecular replacement with existing STAT structures as search models.
- Dimer Interface Analysis: Identify interaction surfaces between β-barrel and four-helix bundle domains using contact analysis software and calculate buried surface areas.
Fluorescence Resonance Energy Transfer (FRET):
- Construct Design: Create STAT fusion proteins with donor (CFP) and acceptor (YFP) fluorophores at positions that report dimerization status without disrupting native structure.
- Live Cell Imaging: Transfert constructs into appropriate cell lines (e.g., HEK293, mouse embryonic fibroblasts) and image using confocal microscopy with appropriate filter sets.
- Quantitative Analysis: Calculate FRET efficiency using acceptor photobleaching or sensitized emission methods to demonstrate separation of dimeric arrangement upon activation [53].
DNA Binding and Gene Regulation Assays
Atomic Force Microscopy (AFM) for DNA-Protein Interactions:
- Sample Preparation: Incubate purified U-STAT proteins with specific DNA fragments containing GAS elements or other target sequences.
- Imaging Conditions: Deposit complexes onto freshly cleaved mica surfaces and image using tapping mode in liquid to preserve native structure.
- Data Analysis: Measure complex dimensions and stoichiometry to determine whether U-STATs bind as dimers or monomers [4].
Electrophoretic Mobility Shift Assay (EMSA):
- Protocol: Incubate purified U-STAT core proteins with ³²P-labeled DNA probes containing high-affinity STAT target sequences.
- Binding Conditions: Include non-specific competitors (poly dI-dC) and specific competitors (unlabeled probe) to demonstrate binding specificity.
- Cysteine Mutation Analysis: Introduce C367A and C542A mutations to test disulfide bridge importance for DNA binding [4].
Chromatin Immunoprecipitation (ChIP) Sequencing:
- Crosslinking: Treat cells with formaldehyde to crosslink proteins to DNA.
- Immunoprecipitation: Use antibodies specific to unphosphorylated STATs (validate phosphorylation state specificity).
- Sequencing and Analysis: Sequence precipitated DNA fragments and map to genome to identify U-STAT-specific binding sites distinct from pSTAT sites [49].
Figure 2: Experimental Workflow for Investigating Unphosphorylated STAT Dimerization and Function. Comprehensive approach combining structural, cellular, and molecular techniques to elucidate non-canonical STAT mechanisms.
Research Reagent Solutions
Table 3: Essential Research Reagents for Studying Unphosphorylated STAT Functions
| Reagent Category | Specific Examples | Research Application |
|---|---|---|
| STAT Expression Constructs | STAT5a ΔNTD ΔTAD; STAT3 Y705F phospho-mutant; STAT1 truncation variants [53] [4] | Structural studies and functional characterization of phosphorylation-independent functions |
| Cell Lines | STAT3 null mouse embryonic fibroblasts; Drosophila hematopoietic tumor models [54] [4] | Reconstitution studies and genetic screening for modifiers |
| Specialized Antibodies | Phospho-specific STAT antibodies; U-STAT specific antibodies [4] [49] | Differentiation between phosphorylation states in immunodetection |
| SH2 Domain Mutants | Double alanine substitutions in Stat6 SH2 domain; Cysteine mutations (C367A/C542A) in STAT3 [51] [4] | Functional analysis of specific residues in dimerization and DNA binding |
| Importin Reagents | Importin-α3 specific inhibitors; Importin binding domain mutants [4] | Nuclear transport mechanism studies |
Implications for Disease and Therapeutic Development
The recognition of non-canonical STAT functions has profound implications for understanding disease pathogenesis and developing targeted therapeutic interventions. Unphosphorylated STATs contribute to various pathological conditions through mechanisms distinct from canonical signaling:
- Cancer pathogenesis: U-STAT3 drives expression of oncogenes and regulates chromatin organization, contributing to malignancy through phosphorylation-independent mechanisms [4].
- Immune dysregulation: Altered U-STAT functions disrupt normal immune cell development and function, potentially contributing to autoimmune and inflammatory conditions [49].
- Therapeutic resistance: Non-canonical STAT signaling may maintain pro-survival gene expression even when canonical signaling is pharmacologically inhibited.
The SH2 domain remains a compelling drug target despite its different role in non-canonical signaling. Developing SH2 domain inhibitors that disrupt both canonical and non-canonical functions could provide more complete pathway suppression than JAK inhibitors alone [15]. Additionally, targeting the specific nuclear import mechanisms of U-STATs or their unique DNA-binding interfaces may enable more precise modulation of non-canonical functions with reduced off-target effects.
Future research directions should focus on developing specific inhibitors of U-STAT dimerization, nuclear transport, and DNA binding, as well as generating more sophisticated experimental models that accurately distinguish between canonical and non-canonical STAT functions in disease contexts.
Liquid-liquid phase separation (LLPS) has emerged as a fundamental mechanism organizing cellular biochemistry, particularly in signal transduction. This technical guide explores the cutting-edge methodologies being deployed to investigate how Src homology 2 (SH2) domains, through their specific recognition of phosphotyrosine motifs, drive the formation of biomolecular condensates in cellular signaling pathways. With a specific focus on STAT (Signal Transducers and Activators of Transcription) proteins, we detail experimental frameworks that bridge molecular biophysics with functional cell biology, providing researchers with robust tools to decipher the role of phase separation in SH2-mediated signaling complexes and its implications for therapeutic intervention.
SH2 domains are modular protein domains of approximately 100 amino acids that specifically recognize and bind to phosphorylated tyrosine (pY) residues, thereby orchestrating a vast network of protein-protein interactions in signal transduction [16] [8]. The human proteome encodes roughly 110 SH2 domain-containing proteins, which are functionally diverse and include enzymes, adaptors, and transcription factors like the STAT family [8]. The canonical function of SH2 domains is to recruit their host proteins to specific pY sites on activated receptors or scaffold proteins, facilitating the assembly of multi-protein signaling complexes.
Recent advances have revealed that beyond simple binary interactions, SH2 domains participate in multivalent interactions that can drive the formation of biomolecular condensates via LLPS [55] [8]. These condensates are membraneless organelles that concentrate specific proteins and nucleic acids, enhancing biochemical reaction rates and providing spatial regulation of signaling [56] [57]. For STAT transcription factors, SH2 domain-mediated dimerization, which is critical for their nuclear function and DNA binding, is now understood to occur within the context of these phase-separated environments [32]. This paradigm shift necessitates new technical approaches to study SH2 domain function, moving from traditional biochemistry to techniques that can capture the dynamics and material properties of these condensed phases. The following sections detail the key experimental strategies for investigating this phenomenon.
Core Techniques for Analyzing SH2-Mediated Condensates
Biochemical Reconstitution and Visualization
A foundational approach for establishing that a protein or complex undergoes LLPS is in vitro reconstitution using purified components. This method allows researchers to control variables such as protein concentration, pH, salt, and the presence of binding partners like lipids or nucleic acids.
Protocol: Minimal Reconstitution of SH2-Mediated Condensates
- Protein Purification: Express and purify recombinant SH2 domain-containing proteins (e.g., full-length STATs, LAT, GRB2) and their cognate binding partners (e.g., phosphorylated peptides or proteins) from systems like E. coli or insect cells [55].
- Droplet Formation Assay: Combine the purified proteins in a physiological buffer. A typical reaction might include:
- SH2 domain-containing protein (e.g., PLCγ1) at a concentration near its known dissociation constant.
- A tyrosine-phosphorylated peptide or protein (e.g., phosphorylated LAT).
- A crowding agent like PEG to mimic the intracellular environment.
- Visualization: Incubate the mixture and visualize droplet formation using differential interference contrast (DIC) or confocal microscopy if the proteins are fluorescently tagged [56] [58].
- Turbidity Assay: Quantify condensate formation by measuring the optical density at 600 nm (OD600). A sharp increase in turbidity indicates the formation of light-scattering droplets [58].
Key Parameters to Test:
- Concentration Dependence: Systematically vary the concentration of the SH2-containing protein and its phospho-ligand to determine the threshold concentration for phase separation [57].
- Environmental Sensitivity: Test the effects of 1,6-hexanediol (an aliphatic alcohol that disrupts hydrophobic interactions), salt concentration, and temperature on condensate stability [56] [58].
Characterizing Material Properties and Dynamics
Once condensates are formed, their material properties—whether liquid-like, gel-like, or solid—must be characterized, as these states have profound functional implications.
Table 1: Techniques for Characterizing Condensate Material Properties
| Technique | Principle | Application in SH2 Signaling | Key Readout |
|---|---|---|---|
| Fluorescence Recovery After Photobleaching (FRAP) | A laser bleaches fluorescence in a region of the condensate; recovery rate indicates internal dynamics and exchange with the surroundings [56] [57]. | Used to demonstrate the liquid nature of LAT/Grb2/PLCγ1 condensates in T-cell receptor signaling [55]. | Half-time of recovery and mobile fraction. Liquid droplets typically show rapid recovery. |
| Fluorescence Correlation Spectroscopy (FCS) | Measures concentration and diffusion rates of fluorescent molecules within condensates and the dilute phase [58]. | Can quantify the enrichment of SH2-domain proteins and their ligands within the condensed phase. | Diffusion coefficients and particle brightness. |
| Inverse Capillary Velocity / Microrheology | Uses the fusion kinetics of two droplets or the motion of embedded particles to measure viscosity and surface tension [58]. | Determines the material state of condensates formed by oncogenic SH2-domain mutants like SHP2 [57]. | Viscosity, surface tension. |
For STAT proteins, a transition from a liquid-like to a more solid, amyloid-like state in pathological conditions can be detected by a drastic slowdown in FRAP recovery [57].
Proximity to Membranes and Phosphatase Protection
SH2-mediated signaling often occurs at the plasma membrane. Reconstitution on supported lipid bilayers (SLBs) provides a more physiologically relevant context.
Protocol: Reconstitution on Supported Lipid Bilayers (SLBs)
- SLB Formation: Create a planar lipid bilayer containing phosphoinositides such as PIP2 or PIP3, which are known to interact with many SH2 domains [8].
- Protein Incubation: Introduce the tyrosine-phosphorylated scaffold protein (e.g., LAT) and the SH2-containing effector proteins (e.g., GRB2, GADS, PLCγ1) to the SLB.
- Imaging: Use Total Internal Reflection Fluorescence (TIRF) microscopy to visualize the formation of membrane-associated condensates with high signal-to-noise ratio [55].
Assay: Phosphatase Protection The dense environment of condensates can shield phosphotyrosine sites from phosphatases. This can be tested by:
- Forming phosphotyrosine-dependent condensates on SLBs.
- Adding a tyrosine phosphatase (e.g., CD45).
- Monitoring the dissociation kinetics of SH2-domain proteins via TIRF. Slower dissociation in condensates indicates protection from dephosphorylation [55].
Advanced and Integrated Methodologies
In Vivo Manipulation with Optogenetics
To move from in vitro observations to functional validation in living cells, optogenetic tools are indispensable. These systems allow spatiotemporal control over LLPS.
Protocol: OptoDroplet System
- Construct Design: Fuse the SH2 domain and/or its flanking intrinsically disordered regions (IDRs) from a protein like STAT to the Cry2 oligomerization domain from Arabidopsis thaliana, which clusters upon exposure to blue light [56] [58].
- Cell Transfection: Introduce the construct into an appropriate cell line.
- Light Induction: Expose cells to blue light to induce rapid and reversible clustering of the fusion protein, mimicking LLPS.
- Functional Assay: Assess downstream consequences, such as the transcription of STAT target genes, to link phase separation to biological function [56].
Computational Prediction and Analysis
Table 2: Computational Tools for Predicting LLPS Propensity
| Tool / Database | Function | Utility for SH2 Protein Research |
|---|---|---|
| D2P2 | Database for disorder predictions and domain annotations [56]. | Identifies intrinsically disordered regions (IDRs) in SH2-containing proteins like STATs that may drive LLPS. |
| PhaSePro | Database of experimentally verified LLPS proteins. | Provides reference data on known drivers and regulators of condensates. |
| DISOPRED3 | Predicts protein disorder from amino acid sequence. | Helps prioritize SH2-domain proteins for LLPS studies based on the presence of flanking disordered regions. |
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Studying SH2-Mediated Phase Separation
| Reagent / Tool | Function / Mechanism | Example Application in SH2 Signaling |
|---|---|---|
| 1,6-Hexanediol | Disrupts hydrophobic interactions; dissolves liquid-like condensates. | Testing the liquidity of LAT/Grb2 condensates; distinguishes liquid from solid aggregates [58] [57]. |
| OptoDroplet System (Cry2) | Blue-light-induced oligomerization to nucleate condensates in vivo. | Studying the functional impact of STAT condensation on gene expression [56] [58]. |
| Supported Lipid Bilayers (SLBs) | In vitro membrane mimetic incorporating PIP2/PIP3. | Reconstituting membrane-proximal condensates in TCR signaling [55] [8]. |
| Phosphatase Inhibitors/Enzymes | Modulate phosphorylation status of scaffold proteins. | Determining the phosphotyrosine-dependence of condensate formation and stability (e.g., using CD45) [55]. |
| Fluorescent Tags (eGFP, mCherry) | Labeling proteins for visualization and FRAP. | Tagging STAT or other SH2-proteins to monitor their dynamics in condensates [57] [55]. |
Visualizing Signaling Pathways and Experimental Workflows
SH2 Domain-Driven Condensates in STAT Activation
Diagram 1: STAT activation pathway. The pathway begins with cytokine-induced receptor activation and phosphorylation, leading to STAT recruitment via its SH2 domain. Subsequent STAT phosphorylation enables reciprocal SH2-pY mediated dimerization, a key step that can occur within biomolecular condensates before the dimer translocates to the nucleus to drive transcription within phase-separated hubs.
Experimental Workflow for LLPS Analysis
Diagram 2: LLPS analysis workflow. A comprehensive experimental pipeline begins with computational prediction of phase separation propensity, proceeds through in vitro reconstitution and biophysical characterization, incorporates membrane context, and culminates in cellular validation and functional assessment.
The integration of LLPS into the framework of SH2-mediated signaling represents a fundamental shift in our understanding of cellular communication. For STAT proteins, the reciprocal SH2-pTyr interaction that underpins dimerization is no longer just a binary switch but a multivalent interaction that can be amplified and regulated within the context of a biomolecular condensate. The techniques outlined here—from quantitative in vitro reconstitution to optogenetic manipulation in cells—provide a roadmap for researchers to dissect the mechanisms and functional consequences of this phenomenon.
The therapeutic implications are substantial. Aberrant phase separation driven by mutant SH2 domain-containing proteins is increasingly linked to cancer and other diseases [57] [8]. The methods described will be crucial for identifying and validating novel therapeutic strategies that target the formation or material properties of pathogenic condensates, potentially leading to a new class of drugs that modulate cell signaling not by inhibiting a single kinase, but by reprogramming the physical organization of the signaling network itself.
Dysregulation and Disease: When STAT-SH2 Dimerization Goes Awry
Autosomal-dominant hyper-IgE syndrome (AD-HIES) provides a compelling model for understanding the profound biological consequences of loss-of-function mutations in a critical signaling pathway. The majority of AD-HIES cases are caused by heterozygous, dominant-negative mutations in the Signal Transducer and Activator of Transcription 3 (STAT3) gene, which disrupt the function of its Src homology 2 (SH2) domain [59] [60]. This domain is indispensable for STAT3 activation and dimerization, processes fundamental to its role as a transcription factor. The SH2 domain facilitates both the recruitment of STAT3 to activated cytokine receptors and the subsequent reciprocal phosphotyrosine-SH2 interactions that stabilize STAT3 dimers for nuclear translocation [5] [32]. In AD-HIES, mutated STAT3 proteins interfere with normal STAT3 function, leading to a multisystem disorder characterized by recurrent infections, elevated serum IgE, eczema, and distinctive connective tissue, skeletal, and vascular abnormalities [59] [60]. This whitepaper examines how STAT3 SH2 domain mutations disrupt normal dimerization and signaling, linking molecular pathology to clinical disease and outlining essential research methodologies for its investigation.
Molecular Basis: The Critical Role of the STAT3 SH2 Domain
Structural and Functional Organization of STAT3
STAT3 is a multifunctional transcription factor composed of several conserved domains: an N-terminal domain (NTD), a coiled-coil domain (CCD), a DNA-binding domain (DBD), a linker domain (LD), an SH2 domain, and a C-terminal transactivation domain (TAD) [4] [10]. The SH2 domain is the central mediator of STAT3 activation. Its primary functions include:
- Receptor Docking: Binding to phosphotyrosine motifs on cytokine receptor complexes.
- Dimerization Stabilization: Mediating reciprocal phosphotyrosine-SH2 interactions between two STAT3 monomers to form active, parallel dimers [5] [32].
- Nuclear Import: Facilitating the nuclear translocation of activated dimers to regulate target gene expression [4].
The canonical activation pathway begins when cytokines (e.g., IL-6, IL-10, IL-21, IL-23) bind to their cognate receptors, triggering JAK kinase-mediated tyrosine phosphorylation of the receptor cytoplasmic tails [33]. STAT3 monomers are recruited to these phosphotyrosine sites via their SH2 domains. Subsequent phosphorylation of STAT3 at tyrosine 705 (Y705) induces a conformational change, enabling SH2-phosphotyrosine reciprocal interaction between two STAT3 monomers to form a stable, parallel dimer [5] [32] [10]. These active dimers translocate to the nucleus, bind gamma-activated sequence (GAS) elements in target gene promoters, and initiate transcription.
Unique Features of the STAT-Type SH2 Domain
STAT-type SH2 domains possess distinctive characteristics that set them apart from Src-type SH2 domains. Structurally, they maintain a central anti-parallel β-sheet (βB-βD strands) flanked by two α-helices (αA and αB), forming an αβββα motif [5]. The STAT-type SH2 domain is uniquely characterized by a C-terminal α-helix (αB'), whereas the Src-type features a β-sheet in this region [5]. This domain contains two crucial sub-pockets:
- pY Pocket (Phosphate-Binding Pocket): Formed by the αA helix, BC loop, and one face of the central β-sheet, this pocket interacts with the phosphotyrosine (pY) residue.
- pY+3 Pocket (Specificity Pocket): Created by the opposite face of the β-sheet, αB helix, and CD and BC* loops, this pocket accommodates residues C-terminal to the pY and confers binding specificity [5].
The SH2 domain also contains a hydrophobic system at the base of the pY+3 pocket that stabilizes the β-sheet and maintains overall domain integrity [5]. Furthermore, the αB, αB', and BC* loop participate in SH2-mediated STAT dimerization, forming critical cross-domain interactions. This structural arrangement means that residues in the pY+3 pocket can exert dual effects on both STAT dimerization capacity and phosphopeptide binding [5].
Figure 1: Canonical STAT3 Activation Pathway. The SH2 domain mediates critical steps in receptor recruitment and dimer stabilization through reciprocal phosphotyrosine interactions.
AD-HIES-Associated STAT3 SH2 Domain Mutations
Mutation Spectrum and Clinical Correlations
Sequencing analyses of patient samples have identified the SH2 domain as a hotspot in the mutational landscape of STAT3, with the majority of AD-HIES cases resulting from heterozygous, dominant-negative mutations in this domain [5] [59]. These mutations are distributed throughout the SH2 domain, affecting residues critical for phosphotyrosine binding, domain stability, and dimerization.
Table 1: Characterized STAT3 SH2 Domain Mutations in AD-HIES
| Mutation | Domain Location | Structural Impact | Clinical Presentation | Reference |
|---|---|---|---|---|
| K591E/M | αA2 helix, pY pocket | Disrupts phosphotyrosine binding | AD-HIES (Germline) | [5] |
| R609G | βB5 strand, pY pocket | Affects Sheinerman & Signature motifs | AD-HIES (Germline) | [5] |
| S611N/G/I | βB7 strand, pY pocket | Disrupts Sheinerman & Signature motifs | AD-HIES (Germline) | [5] |
| S614R | BC loop, pY pocket | Alters phosphotyrosine binding | T-LGLL, NK-LGLL (Somatic) | [5] |
| E616G/K | BC loop, pY pocket | Disrupts BC loop structure | DLBCL, NKTL (Somatic) | [5] |
| G617E/V/R | BC loop, pY pocket | Impairs BC loop conformation | AD-HIES (Germline) | [5] |
Notably, the genetic volatility of specific regions in the SH2 domain can result in either activating or deactivating mutations at the same site, underscoring the delicate evolutionary balance of wild-type STAT structural motifs in maintaining precise levels of cellular activity [5]. For instance, while the S614R mutation is predominantly found in somatic cases of T-cell large granular lymphocytic leukemia (T-LGLL) and natural killer (NK)-LGLL, different mutations at the same residue can lead to varied clinical outcomes [5].
Mechanisms of Dominant-Negative Interference
AD-HIES-associated STAT3 mutations typically follow a dominant-negative (DN) pattern of inheritance, where the mutant STAT3 allele interferes with the function of the wild-type allele. Approximately 95% of in-frame STAT3 variants demonstrate dominant-negative effects [59]. The molecular mechanisms include:
- Aberrant Dimer Formation: Mutant STAT3 can dimerize with wild-type STAT3, but these dimers are dysfunctional and cannot bind DNA effectively or initiate transcription.
- Competitive Receptor Binding: Mutant STAT3 competes with wild-type STAT3 for docking sites on activated cytokine receptors, sequestering recruitment sites.
- Disruption of Nuclear Translocation: Improperly formed dimers fail to undergo nuclear import, despite sometimes retaining phosphorylation capacity [61].
The dominant-negative effect explains why heterozygous mutations cause disease despite the presence of a functional wild-type allele. This effect is particularly impactful in STAT3 signaling, where precise dimer stoichiometry is crucial for proper transcriptional regulation of target genes.
Immunological and Clinical Consequences of STAT3 Dysfunction
Pathway-Specific Signaling Deficits
The disruption of STAT3 dimerization and nuclear translocation in AD-HIES leads to profound, pathway-specific signaling deficits. STAT3 is activated by numerous cytokines, including IL-6, IL-10, IL-21, IL-22, and IL-23, which signal through receptors containing the gp130 subunit or the common gamma chain (γc) [59] [33]. The immunological consequences are particularly evident in several key pathways:
- Th17 Cell Deficiency: Impaired IL-6, IL-21, and IL-23 signaling disrupts the differentiation of T helper 17 (Th17) cells, which are essential for mucosal immunity against fungi and extracellular bacteria [59] [60]. AD-HIES patients typically have markedly reduced Th17 cells (less than 0.3% of CD4+ T cells), explaining their susceptibility to candidiasis and staphylococcal infections [59].
- Impaired IL-10 Signaling: IL-10 exerts potent anti-inflammatory effects through STAT3 activation. In AD-HIES, IL-10 signaling is defective in monocytes and dendritic cells, leading to reduced expression of anti-inflammatory mediators like IL-1 receptor antagonist (IL-1ra) and SOCS3, and enhanced production of pro-inflammatory cytokines such as TNF-α, IL-6, and IL-12 [62]. This creates an imbalance between pro-inflammatory and anti-inflammatory signals, contributing to persistent inflammation and delayed healing after infections [62].
- B Cell Dysregulation: STAT3 is crucial for naïve B cell activation, affinity maturation, and class switching. Patients exhibit impaired antigen-specific antibody responses and decreased memory B cell levels, despite normal or elevated total immunoglobulin levels (except IgE) [59].
Table 2: Key Signaling Deficits in AD-HIES and Their Clinical Correlates
| Deficient Pathway | Affected Cell Types | Functional Consequences | Clinical Manifestations |
|---|---|---|---|
| IL-6/IL-23 Signaling | CD4+ T cells | Reduced Th17 differentiation | Mucocutaneous candidiasis, recurrent staphylococcal abscesses |
| IL-10 Signaling | Monocytes, Neutrophils, DCs | Excessive pro-inflammatory cytokine production | Cold abscesses, persistent inflammation |
| IL-21 Signaling | CD8+ T cells, B cells | Impaired cytotoxic T cell function, reduced memory B cells | Severe viral infections, impaired vaccine responses |
| IL-11/IL-6 Signaling | Osteoblasts, Osteoclasts | Skeletal homeostasis disruption | Osteopenia, minimal trauma fractures |
| LIF Signaling | Mesenchymal cells | Connective tissue regulation | Scoliosis, abnormal facies, retained primary teeth |
Broad Clinical Phenotype of AD-HIES
The clinical manifestations of AD-HIES extend beyond immunodeficiency to include multisystem abnormalities, reflecting the widespread role of STAT3 in development and homeostasis:
- Infectious Phenotype: Patients suffer from recurrent staphylococcal skin abscesses ("cold" abscesses without warmth or erythema), recurrent pneumonias (often leading to pneumatocele formation), and chronic mucocutaneous candidiasis [60].
- Immunological Findings: Extremely elevated serum IgE levels (>2000 IU/mL) and peripheral eosinophilia are hallmark features, though the exact mechanistic link to STAT3 dysfunction remains incompletely understood [60].
- Non-Immune Manifestations: These include characteristic facial features (asymmetric facies, prominent forehead, wide-set eyes), skeletal abnormalities (scoliosis, osteopenia, minimal trauma fractures), retained primary teeth, and vascular abnormalities (coronary artery aneurysms, lacunar infarcts) [60].
The NIH HIES scoring system incorporates many of these features to aid diagnosis, with most STAT3-deficient patients scoring ≥40 points [59]. However, the clinical spectrum continues to expand with the identification of atypical cases displaying mixed phenotypes that challenge the simple loss-of-function versus gain-of-function dualism [61].
Experimental Approaches for Investigating STAT3 SH2 Domain Mutations
Biosensor Technologies for Monitoring STAT Dimerization
Advanced biosensor technologies have revolutionized the study of STAT activation dynamics. STATeLights represent a class of genetically encoded biosensors that enable direct, continuous detection of STAT activity in live cells with high spatiotemporal resolution [10]. These biosensors utilize fluorescence lifetime imaging microscopy-Förster resonance energy transfer (FLIM-FRET) to monitor conformational changes during STAT dimerization.
The STATeLight biosensor engineering strategy involves:
- Fluorescent Protein Tagging: Tagging STAT monomers with a FRET pair (mNeonGreen donor and mScarlet-I acceptor) at strategic positions.
- Conformational Sensing: Optimizing fusion sites to detect the cytokine-induced conformational change from antiparallel to parallel dimers.
- FLIM-FRET Detection: Measuring FRET efficiency changes via fluorescence lifetime, which is inversely correlated with FRET efficiency [10].
The most effective STAT5A biosensor (variant 4) involved C-terminal fusion of fluorophores to truncated STAT5A containing the core fragment, resulting in up to 12% FRET efficiency upon IL-2 stimulation [10]. This approach directly monitors conformational rearrangement of STAT dimers and is unaffected by inactive phosphorylated monomers or truncated variants, providing specific observation of STAT activation.
Protocol: Investigating STAT3 Dimerization via FRET/FLIM
Objective: To quantitatively assess STAT3 dimerization dynamics in live cells expressing wild-type or SH2 domain mutant STAT3.
Materials:
- HEK-Blue IL-2 cells (or similar cytokine-responsive cell line)
- STATeLight biosensor constructs (donor and acceptor fusions)
- Recombinant cytokines (IL-6, IL-10, IL-21)
- FLIM-capable confocal microscope system
- Transfection reagents
Methodology:
- Cell Transfection: Transfect cells with STATeLight biosensor constructs (STAT3 tagged with mNeonGreen and mScarlet-I at C-termini).
- Stimulation: Treat cells with STAT3-activating cytokines (e.g., IL-6 at 50 ng/mL) for varying durations (0-60 minutes).
- FLIM Imaging: Acquire fluorescence lifetime images of mNeonGreen using multiphoton excitation (e.g., 950 nm) and time-correlated single-photon counting.
- Data Analysis:
- Calculate fluorescence lifetime values per pixel.
- Generate lifetime histograms and determine mean lifetime values.
- Compute FRET efficiency using the formula:
E = 1 - (τ_DA/τ_D), where τDA is donor lifetime in presence of acceptor, and τD is donor lifetime alone.
- Validation: Compare dimerization kinetics of wild-type versus mutant STAT3 under identical stimulation conditions [10].
This protocol enables real-time tracking of STAT3 activation, facilitating quantitative comparison of dimerization efficiency between wild-type and disease-associated STAT3 mutants.
Figure 2: Experimental Workflow for STAT3 Dimerization Analysis. The FRET/FLIM-based approach enables real-time monitoring of dimerization dynamics in live cells.
Additional Key Methodologies
Several complementary approaches are essential for comprehensive characterization of STAT3 SH2 domain mutations:
- Phospho-STAT Flow Cytometry: Intracellular staining with phospho-specific STAT3 (pY705) antibodies following cytokine stimulation provides a semi-quantitative measure of STAT3 activation, though it requires cell fixation and permeabilization [10].
- Electrophoretic Mobility Shift Assays (EMSAs): Assess DNA-binding capacity of STAT3 dimers to GAS elements, revealing functional competence of dimeric complexes [4].
- Co-Immunoprecipitation Studies: Evaluate protein-protein interactions between wild-type and mutant STAT3 proteins, demonstrating dominant-negative dimer formation [61].
- Transcriptional Reporter Assays: Measure STAT3-driven luciferase activity under control of GAS-containing promoters to quantify functional output of signaling pathways [61].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for STAT-SH2 Domain Investigation
| Reagent/Category | Specific Examples | Research Application | Key Features & Considerations |
|---|---|---|---|
| STAT Biosensors | STATeLights (FRET/FLIM) | Live-cell dimerization kinetics | High spatiotemporal resolution; requires specialized imaging systems [10] |
| Phospho-Specific Antibodies | anti-STAT3 (pY705) | Detection of STAT3 activation | Requires cell fixation; semi-quantitative [10] |
| Cytokine Reagents | Recombinant IL-6, IL-10, IL-21, IL-23 | Pathway-specific stimulation | Quality and bioactivity vary between suppliers; concentration optimization needed |
| Cell Line Models | HEK-Blue IL-2, STAT3-deficient lines | Controlled genetic background | Ensure cytokine responsiveness; verify STAT3 expression status |
| Expression Vectors | Wild-type/mutant STAT3 constructs | Structure-function studies | Include proper tagging for detection; consider expression levels |
| JAK/STAT Inhibitors | JAK inhibitors (Ruxolitinib) | Pathway modulation controls | Assess specificity; potential off-target effects |
The study of loss-of-function STAT3 mutations in AD-HIES provides profound insights into the critical role of the SH2 domain in STAT dimerization and cellular signaling. The molecular pathology of AD-HIES demonstrates how specific disruptions in SH2 domain function impair dimer stabilization, nuclear translocation, and transcriptional activity, leading to a complex multisystem disorder. Research in this area continues to evolve beyond the simple loss-of-function/gain-of-function paradigm, with emerging evidence of "confusion-of-function" mutations that display mixed behaviors in vitro and ambiguous clinical phenotypes [61].
Future research directions should focus on:
- Developing targeted therapeutic approaches that bypass the defective STAT3 dimerization process
- Elucidating structure-function relationships of atypical STAT3 mutations
- Exploring gene correction strategies for severe cases
- Investigating the role of unphosphorylated STAT3 (U-STAT3) in disease pathogenesis [4]
The experimental approaches outlined in this whitepaper, particularly real-time biosensor technologies, provide powerful tools to advance these investigations and develop targeted interventions for STAT3-related diseases.
Signal Transducer and Activator of Transcription (STAT) proteins represent a critical family of transcription factors that mediate cellular responses to cytokines, growth factors, and hormones. The Src Homology 2 (SH2) domain within STAT proteins serves as a fundamental molecular switch, governing activation through phosphotyrosine recognition and subsequent dimerization. In normal physiology, STAT activation is transient and tightly regulated; however, genetic alterations disrupting this precision can yield gain-of-function (GOF) mutants that drive oncogenesis. Research has revealed that the SH2 domain is a mutational hotspot in STAT proteins, particularly in hematologic malignancies, where these mutations can lead to either hyperactivated or refractory STAT mutants with profound pathological consequences [5]. Understanding the structural and functional implications of these mutations provides the foundation for targeted therapeutic interventions in leukemias and lymphomas, framing a critical research domain that intersects molecular oncology and drug discovery.
The centrality of the STAT-SH2 domain in malignant transformation stems from its dual role in mediating recruitment to activated cytokine receptors and facilitating STAT dimerization—a prerequisite for nuclear translocation and transcriptional activity. Sequencing analyses of patient samples have consistently identified the SH2 domain as a hotspot in the mutational landscape of STAT proteins, particularly STAT3 and STAT5B, although the functional impact for the vast majority of these mutations remains poorly characterized [5]. This review synthesizes current understanding of how GOF mutations within the STAT-SH2 domain contribute to oncogenic activation, with particular emphasis on their roles in leukemias and lymphomas, while providing detailed methodological frameworks for their investigation.
Molecular Architecture of STAT-SH2 Domains and Dysregulation by Mutation
Structural Basis of STAT-SH2 Domain Function
SH2 domains are modular protein interaction domains approximately 100 amino acids in length that specifically recognize phosphorylated tyrosine motifs. The STAT-type SH2 domain exhibits a characteristic αβββα fold consisting of a central anti-parallel β-sheet (βB-βD strands) flanked by two α-helices (αA and αB) [5] [8]. This structure forms two functionally critical subpockets: the phosphotyrosine (pY) binding pocket and the pY+3 specificity pocket. The pY pocket, formed by the αA helix, BC loop, and one face of the central β-sheet, contains a highly conserved arginine residue (within the FLVR motif) that directly coordinates the phosphate group of phosphotyrosine through a salt bridge [16] [8]. The pY+3 pocket, created by the opposite face of the β-sheet along with residues from the αB helix and CD and BC* loops, determines binding specificity by accommodating residues C-terminal to the phosphotyrosine [5].
STAT-type SH2 domains are distinguished from Src-type SH2 domains by the presence of an additional α-helix (αB') at the C-terminal region instead of β-sheets, an adaptation that facilitates STAT dimerization—a critical step in STAT-mediated transcriptional regulation [8]. This structural specialization enables the reciprocal SH2-phosphotyrosine interactions that form active STAT dimers following phosphorylation. Conventional phosphopeptide binding occurs perpendicular to the β-sheet, with the phosphotyrosine interacting with conserved residues in the pY pocket while C-terminal residues extend across the SH2 domain into the pY+3 pocket [5]. These interactions are crucial for maintaining proper binding to facilitate protein dimerization, and specific mutations in these regions can profoundly alter normal STAT function.
Mechanisms of SH2 Domain Mutational Disruption
Mutations within the STAT-SH2 domain can disrupt normal function through multiple mechanisms, with the specific structural location often determining the pathological consequence. The genetic volatility of specific regions in the SH2 domain can result in either activating or deactivating mutations at the same site, underscoring the delicate evolutionary balance of wild-type STAT structural motifs in maintaining precise levels of cellular activity [5]. Several key mutational mechanisms have been identified:
Dimerization Stabilization: Mutations such as STAT5B N642H (located in the SH2 domain) enhance STAT dimer stability by strengthening reciprocal SH2-phosphotyrosine interactions between monomers. This results in prolonged phosphorylation status and nuclear retention, effectively amplifying transcriptional signals [63]. Experimental evidence demonstrates that while pSTAT expression disappears within 1 hour after transient IL-2 stimulation in wild-type STAT5B transduced cells, it persists for more than 6 hours in STAT5B N642H mutant-transduced cells [63].
Phosphopeptide Binding Alteration: Mutations within the pY or pY+3 pockets can modify binding affinity for phosphotyrosine motifs, either enhancing or diminishing recruitment to receptor complexes. For instance, mutations at the conserved arginine residue in the FLVR motif fundamentally disrupt phosphotyrosine recognition, while alterations in the specificity pocket can change receptor binding preferences [5] [8].
Conformational Shifts: Substitutions at critical positions can induce allosteric changes that stabilize active configurations or disrupt autoinhibitory interactions. Protein flexibility is a particular consideration, as STAT SH2 domains exhibit particularly dynamic behavior even in sub-microsecond timescales, with the accessible volume of the pY pocket varying dramatically [5].
Table 1: Classification of STAT-SH2 Domain Mutations by Mechanism and Pathogenic Outcome
| Mutation Type | Molecular Mechanism | Pathogenic Effect | Representative Examples |
|---|---|---|---|
| Dimerization-Stabilizing | Enhanced SH2-pTyr interaction between STAT monomers | Prolonged activation, nuclear retention, amplified transcription | STAT5B N642H, STAT3 S614R |
| Binding Pocket Disruption | Altered affinity for phosphotyrosine motifs | Aberrant receptor recruitment, modified signaling specificity | STAT3 K591E/M, STAT3 R609G |
| Allosteric Modification | Conformational shifts stabilizing active states | Constitutive activation independent of upstream signals | STAT5B mutations in hydrophobic core |
| Compound Dysfunction | Combined effects on multiple interaction interfaces | Complex gain-of-function phenotypes with pathway crosstalk | Multiple simultaneous SH2 domain mutations |
STAT-SH2 Domain Mutations in Hematologic Malignancies
Prevalence and Spectrum Across Disease Entities
Activating mutations of STAT3 and STAT5B are recurrent findings across a spectrum of T-cell malignancies, with particular prevalence in certain disease entities. These mutations are predominantly focused in the SH2 domain—the region responsible for phosphotyrosine docking, transient binding to cytokine receptors, and mediating dimerization with other STAT proteins [63]. The following hematologic malignancies demonstrate significant frequencies of STAT-SH2 domain mutations:
T-cell Large Granular Lymphocytic Leukemia (T-LGLL): STAT3 mutations represent the most common genetic alteration in T-LGLL, with the SH2 domain serving as the predominant mutational hotspot. The most frequent STAT3 mutation in T-LGLL is Y640F, located in the SH2 domain, which promotes constitutive dimerization and activation [5] [63]. Additional STAT3 mutations in T-LGLL include D661Y/V, N647I, and S614R, all clustering within the SH2 domain and resulting in similar GOF effects [5].
Natural Killer/T-cell Lymphoma (NKTL): Both STAT3 and STAT5B SH2 domain mutations are recurrent in NKTL, with STAT5B N642H being particularly prominent. This mutation is located at a residue analogous to STAT3 N647, which is also mutated in T-LGLL, highlighting the functional conservation between different STAT family members [5] [64]. NKTL cases harboring these mutations demonstrate hyperactivation of the JAK-STAT signaling pathway, which drives proliferation and survival of malignant cells [64].
Anaplastic Large Cell Lymphoma (ALCL): STAT3 mutations are observed in both ALK-positive and ALK-negative ALCL variants, with the SH2 domain again representing the primary mutational cluster. These mutations contribute to constitutive STAT3 phosphorylation and subsequent upregulation of anti-apoptotic and proliferative target genes [5] [63].
Other T-cell Malignancies: STAT3 and STAT5B SH2 domain mutations have also been documented in T-cell prolymphocytic leukemia (T-PLL), cutaneous T-cell lymphoma (CTCL), and adult T-cell leukemia/lymphoma (ATLL), albeit at lower frequencies [5] [63].
Table 2: Spectrum of STAT-SH2 Domain Mutations in Hematologic Malignancies
| Malignancy | STAT Gene | Prevalent SH2 Mutations | Reported Frequency | Clinical Associations |
|---|---|---|---|---|
| T-LGLL | STAT3 | Y640F, D661Y/V, N647I, S614R | 30-40% | Chronic neutropenia, autoimmune associations |
| NKTL | STAT5B | N642H | 10-20% | Aggressive disease, EBV association |
| NKTL | STAT3 | Multiple SH2 mutations | 5-15% | Advanced stage, extranodal disease |
| ALCL | STAT3 | D661Y, S614R, others | 10-20% | Potential prognostic significance |
| T-PLL | STAT5B | N642H, others | 5-10% | Aggressive clinical course |
| CTCL | STAT3 | Multiple SH2 mutations | 5-10% | Disease progression |
Functional Consequences of Pathogenic Mutations
The functional impact of STAT-SH2 domain GOF mutations manifests at cellular, pathway, and organismal levels, driving oncogenesis through multiple interconnected mechanisms:
Constitutive Pathway Activation: SH2 domain mutations stabilize STAT dimers independent of physiological activation signals, leading to constitutive nuclear localization and continuous transcription of target genes. This results in persistent activation of transcriptional programs that normally occur only transiently following cytokine stimulation [5] [49]. The structural basis for this constitutive activation involves mutations that enhance the affinity of reciprocal SH2-phosphotyrosine interactions between STAT monomers or that disrupt autoinhibitory regions that maintain STATs in latent configurations.
Transcriptional Reprogramming: Mutant STAT proteins drive expression of genes regulating apoptosis (BCL-2, BCL-XL, MCL-1), cell cycle progression (C-MYC, D-type cyclins), and self-reinforcement (STAT3/STAT5) [5]. This reprogramming creates a cellular state primed for survival and proliferation while resisting normal apoptotic signals. The specific transcriptional profile varies by STAT family member and cellular context but consistently promotes oncogenic transformation.
Cellular Addiction: Malignant cells harboring STAT-SH2 mutations frequently develop dependency on these mutant proteins—a phenomenon termed "oncogene addiction." Loss-of-function studies using shRNA approaches have demonstrated that pSTAT-positive malignant T-cell lines require STAT3 for survival regardless of mutation status, though mutations further enhance this dependency [63]. This addiction creates a therapeutic window potentially exploitable through targeted inhibition.
Ecosystem Interactions: Mutant STAT proteins alter the tumor microenvironment through regulation of cytokine and chemokine expression, influencing immune cell recruitment and function. Additionally, there is growing recognition of non-canonical STAT functions—including roles for unphosphorylated STATs in gene regulation—that may be co-opted or enhanced by SH2 domain mutations [49].
Experimental Approaches for Characterizing STAT-SH2 Domain Mutations
Methodologies for Functional Validation
Comprehensive characterization of STAT-SH2 domain GOF mutations requires integrated experimental approaches spanning biochemical, cellular, and genomic techniques. The following methodologies represent essential components for rigorous functional validation:
Phosphorylation Status Analysis: Assessment of STAT phosphorylation provides fundamental insight into mutation-induced activation. Protocol: Cells are serum-starved, stimulated with relevant cytokines, and lysed. Phosphorylated STAT is detected via western blot using phospho-specific antibodies (e.g., pSTAT3-Y705, pSTAT5-Y694) with total STAT antibodies confirming equal loading. For temporal analysis, phosphorylation persistence is evaluated following cytokine withdrawal, with GOF mutants typically showing prolonged phosphorylation (e.g., >6 hours for STAT5B N642H versus <1 hour for wild-type) [63].
Nuclear Localization Studies: GOF mutations enhance nuclear accumulation of STAT dimers. Protocol: Immunofluorescence staining is performed on fixed cells using STAT-specific antibodies with fluorescent secondary antibodies. DAPI counterstaining identifies nuclei, and confocal microscopy quantifies nuclear-to-cytoplasmic fluorescence ratios. Alternatively, cellular fractionation followed by western blotting provides biochemical confirmation of nuclear translocation [65] [49].
Dimerization Assays: Direct assessment of STAT dimer stability is crucial for SH2 mutant characterization. Protocol: Co-immunoprecipitation of differentially tagged STAT constructs (e.g., FLAG-STAT3 with HA-STAT3) from transfected cells, followed by detection with tag-specific antibodies. Alternatively, native PAGE separates STAT dimers from monomers based on migration mobility, with GOF mutants showing enhanced dimer formation even without cytokine stimulation [5] [49].
Transcriptional Activity Reporter Assays: Functional output of STAT mutants is quantified using luciferase reporters under control of STAT-responsive promoters (e.g., M67-SIE for STAT3, GAS elements for STAT5). Protocol: Cells are co-transfected with STAT mutants and reporter constructs, stimulated with cytokines or left unstimulated, and luciferase activity is measured. GOF mutants typically demonstrate elevated basal activity and/or enhanced cytokine-induced reporter activation compared to wild-type STATs [5].
Gene Expression Profiling: Global transcriptional consequences are assessed via RNA-seq or Nanostring analyses. Protocol: Cells expressing STAT mutants versus wild-type controls are subjected to transcriptome analysis, followed by pathway enrichment assessment. Expected findings include upregulation of anti-apoptotic genes, cell cycle regulators, and inflammatory mediators, with validation of key targets by qRT-PCR [5] [49].
Diagram 1: Experimental Workflow for Characterizing STAT-SH2 Domain Mutations
Essential Research Reagents and Tools
Table 3: Essential Research Reagents for STAT-SH2 Domain Mutation Studies
| Reagent Category | Specific Examples | Experimental Application | Technical Considerations |
|---|---|---|---|
| Expression Plasmids | Wild-type STAT3/5, SH2 mutants (Y640F, N642H, D661Y, etc.) | Functional comparison studies | Tag with FLAG/HA for detection; use lentiviral systems for stable expression |
| Phospho-Specific Antibodies | pSTAT3 (Y705), pSTAT5 (Y694), total STAT3/5 | Activation status assessment | Validate specificity with phosphorylation-deficient mutants |
| Cell Line Models | Ba/F3, HEK293T, NK/T-cell lines (NK-92, YT), patient-derived cells | Context-specific functional assays | Choose models with relevant background (IL-2/7 dependency for T-cell context) |
| JAK/STAT Inhibitors | Tofacitinib (pan-JAK), Ruxolitinib (JAK1/2), PRN371 (JAK3-selective) | Pathway dependency assessment | Consider selectivity profiles; use multiple inhibitors to confirm specificity |
| Reporter Constructs | GAS-luciferase, M67-SIE-luciferase, specific gene promoters | Transcriptional activity measurement | Include mutant GAS elements as negative controls |
| Cytokines/Growth Factors | IL-2, IL-6, IL-21, IFN-γ, EGF | Pathway stimulation | Titrate concentrations for optimal activation without saturation |
Therapeutic Targeting of Oncogenic STAT Signaling
Current and Emerging Intervention Strategies
The pervasive activation of STAT signaling in hematologic malignancies has motivated diverse therapeutic approaches targeting different nodes within the pathway:
JAK Kinase Inhibitors: First-generation JAK inhibitors (tofacitinib, ruxolitinib) demonstrate clinical activity but are limited by their pan-JAK inhibition profiles, which lead to off-target toxicities such as thrombocytopenia (JAK2 inhibition) [63] [64]. Next-generation selective inhibitors (PRN371) specifically target JAK3 through covalent binding to the unique Cys909 residue, achieving 282-fold selectivity over TYK2, 378-fold over JAK2, and 1194-fold over JAK1 [64]. These agents effectively suppress proliferation and induce apoptosis in JAK3-driven malignancies with reduced toxicity.
Direct STAT Inhibitors: Development of direct STAT inhibitors represents an active area of investigation, with particular focus on the SH2 domain due to its critical role in dimerization. Small molecules targeting the SH2 domain aim to disrupt STAT dimerization and nuclear translocation; however, challenges include the relatively shallow binding surfaces and high flexibility of STAT SH2 domains [5] [8]. To date, no direct STAT inhibitors have reached clinical approval, though several candidates are in preclinical development.
Combination Therapies: Given the limitations of monotherapies, rational combinations show enhanced efficacy. For instance, combining JAK1/2 inhibitors with Bcl-xL inhibitors (navitoclax) demonstrates additive/synergistic activity in T-cell malignancy models [63]. This approach simultaneously targets upstream signaling and downstream survival effectors, addressing compensatory resistance mechanisms.
Alternative Modalities: Emerging strategies include proteolysis-targeting chimeras (PROTACs) for directed STAT degradation, protein-protein interaction inhibitors specifically disrupting STAT dimerization, and approaches targeting non-canonical STAT functions [49].
Biomarker Development and Resistance Considerations
Effective translation of STAT-targeted therapies requires robust biomarker strategies and anticipation of resistance mechanisms:
Predictive Biomarkers: The presence of nuclear pSTAT3 or pSTAT5 represents a more reliable biomarker for JAK/STAT pathway activation than mutation status alone, as pathway hyperactivity is more pervasive than specific mutations [63]. Additional biomarkers include gene expression signatures reflecting STAT pathway activation and mutational status of upstream pathway components.
Resistance Mechanisms: Clinical experience reveals several resistance pathways, including activating mutations downstream of JAKs (particularly in STAT proteins themselves), bypass signaling through alternative pathways, and pharmacological limitations such as inadequate target coverage with intermittent dosing [63] [64]. The observation that STAT3-activating mutations can confer resistance to JAK3 inhibitors highlights the challenge of sequential pathway inhibition [64].
Diagram 2: JAK-STAT Signaling Pathway with Oncogenic Perturbations and Therapeutic Interventions
Future Directions and Research Priorities
The investigation of STAT-SH2 domain GOF mutations continues to evolve, with several emerging research priorities shaping future directions:
Structural Dynamics and Allostery: Advanced structural techniques including cryo-electron microscopy and molecular dynamics simulations are needed to elucidate the dynamic changes induced by SH2 mutations. Particular focus should be placed on understanding allosteric networks connecting the SH2 domain to other STAT functional regions, which may reveal novel targeting opportunities [5] [8].
Non-Canonical STAT Functions: Growing evidence indicates that STAT proteins possess functions beyond their canonical role as inducible transcription factors, including gene repression capabilities and roles outside the nucleus mediated by both phosphorylated and unphosphorylated forms [49]. The impact of SH2 mutations on these alternative modalities remains largely unexplored but may significantly contribute to oncogenesis.
Evolutionary Context: STAT-type SH2 domains represent an ancient adaptation predating animal multicellularity, with conserved examples in organisms like Dictyostelium that employ SH2 domain/phosphotyrosine signaling for transcriptional regulation [8]. Understanding the evolutionary constraints on SH2 structure and function may provide insights into mutation vulnerability and compensatory mechanisms.
Microenvironmental Interactions: STAT mutations likely alter tumor cell communication with the immune microenvironment through modified cytokine production and responsiveness. Comprehensive analysis of these ecosystem-level effects may identify immune-modulatory opportunities to enhance antitumor responses.
Clinical Translation Accelerators: Improved preclinical models including patient-derived xenografts and genetically engineered mouse models that faithfully recapitulate human STAT-driven malignancies are needed. Additionally, clinical trial designs should incorporate biomarker-stratified approaches and rational combination strategies based on mechanistic insights into resistance pathways.
In conclusion, STAT-SH2 domain GOF mutations represent a paradigm of oncogenic transformation through transcription factor activation, with particular significance in hematologic malignancies. Their study integrates structural biology, cancer genetics, and therapeutic development, offering a compelling research framework with direct clinical implications. As investigation continues to unravel the complexities of STAT signaling, the potential for precision medicine approaches targeting these pathogenic drivers continues to expand.
The Src Homology 2 (SH2) domain is a critical modular unit that arose approximately 600 million years ago within metazoan signaling pathways, intricately tied to multicellular signal transduction [5] [8]. In STAT (Signal Transducer and Activator of Transcription) proteins, the SH2 domain serves as a master regulator of canonical activation, mediating receptor recruitment, phosphorylation-dependent dimerization, and nuclear translocation of phosphorylated STAT dimers to drive transcription of target genes [5]. The SH2 domain achieves this through a conserved structural fold featuring a central anti-parallel β-sheet flanked by two α-helices, creating distinct phosphate-binding (pY) and specificity (pY+3) pockets that determine phosphopeptide selectivity [5] [8].
Sequencing analyses of patient samples have revealed the SH2 domain as a hotspot in the mutational landscape of STAT proteins, particularly STAT3 and STAT5 [5]. What makes this region extraordinarily compelling is its genetic volatility—specific residues can yield diametrically opposed functional consequences when mutated, creating a delicate evolutionary balance where the same genomic position can produce either gain-of-function (GOF) or loss-of-function (LOF) mutations depending on the specific amino acid substitution [5]. This review examines the structural and mechanistic basis for these opposing functional outcomes within STAT-SH2 domains, exploring implications for disease pathogenesis, experimental investigation, and therapeutic intervention.
Structural Framework of STAT-Type SH2 Domains
Unique Architectural Features of STAT SH2 Domains
STAT-type SH2 domains exhibit distinctive structural characteristics that set them apart from Src-type SH2 domains found in other signaling proteins. While all SH2 domains share a conserved αβββα core motif, STAT-type domains are characterized by:
- C-terminal α-helix (αB') instead of the β-sheet (βE and βF strands) found in Src-type domains [5]
- Shorter CD loops compared to enzymatic SH2 domain-containing proteins [8]
- Specific adaptations for dimerization, reflecting ancestral functions predating animal multicellularity [8]
The STAT SH2 domain contains several functionally critical regions. The pY pocket (phosphate-binding pocket) is formed by the αA helix, BC loop, and one face of the central β-sheet, containing an invariant arginine at position βB5 (part of the FLVR motif) that directly engages phosphotyrosine through salt bridge interactions [5] [8]. The adjacent pY+3 pocket (specificity pocket) is created by the opposite face of the β-sheet along with residues from the αB helix and CD/BC* loops, determining phosphopeptide binding specificity [5]. Additionally, the evolutionary active region (EAR) at the C-terminal portion of the pY+3 pocket and a hydrophobic system of non-polar residues at the base of this pocket provide structural stability and additional functional versatility [5].
SH2 Domain Role in STAT Activation Cycle
The SH2 domain is indispensable for canonical STAT activation, which follows a precisely regulated sequence:
- Cytokine/Growth Factor Binding: Extracellular ligands bind their cognate receptors, activating associated Janus kinases (JAKs) [4]
- Receptor Phosphorylation: JAKs phosphorylate tyrosine residues on receptor intracellular domains [4] [10]
- STAT Recruitment: STAT monomers are recruited to phosphorylated receptors via their SH2 domains [5]
- STAT Phosphorylation: JAKs phosphorylate a conserved tyrosine residue in the STAT C-terminal transactivation domain (Tyr705 in STAT3) [4]
- SH2-Mediated Dimerization: Phosphorylated STATs form parallel homodimers or heterodimers through reciprocal SH2-phosphotyrosine interactions [4] [53]
- Nuclear Translocation: STAT dimers translocate to the nucleus and bind gamma-activated sequence (GAS) elements to regulate transcription [4]
Table 1: Key Structural Motifs in STAT SH2 Domains and Their Functional Roles
| Structural Motif | Location | Key Residues | Functional Role |
|---|---|---|---|
| pY Pocket | αA helix, BC loop, βB strand | Invariant Arg βB5 | Phosphotyrosine binding via salt bridge |
| pY+3 Pocket | Opposite β-sheet face, αB helix, CD/BC* loops | Variable specificity determinants | Phosphopeptide selectivity |
| Evolutionary Active Region (EAR) | C-terminal pY+3 pocket | αB' helix residues | STAT-type specific adaptations |
| Hydrophobic System | Base of pY+3 pocket | Cluster of non-polar residues | Structural integrity maintenance |
| Dimerization Interface | αB, αB', BC* loop | Cross-domain interacting residues | Mediates STAT dimer formation |
Mutational Hotspots with Opposing Functional Outcomes
The Genetic Volatility of Specific SH2 Domain Positions
The STAT SH2 domain represents a remarkable example of how specific positions within a protein can serve as mutational hotspots with opposing functional consequences. Patient sequencing data has identified numerous disease-associated mutations within the STAT3 and STAT5B SH2 domains where the same residue can yield either activating or inactivating mutations depending on the specific amino acid substitution [5].
Table 2: STAT3 SH2 Domain Mutational Hotspots with Opposing Functional Consequences
| Residue Position | Structural Location | LOF Mutations | GOF Mutations | Associated Pathologies |
|---|---|---|---|---|
| Ser611 | βB7 strand (pY pocket) | S611G, S611N, S611I (AD-HIES) | - | Autosomal-dominant Hyper IgE Syndrome (LOF) |
| Ser614 | BC loop (pY pocket) | S614G (AD-HIES) | S614R (Cancer) | T-LGLL, NK-LGLL, ALK-ALCL, HSTL (GOF) |
| Glu616 | BC loop (pY pocket) | - | E616G, E616K (Cancer) | DLBCL, NKTL (GOF) |
| Gly617 | BC loop (pY pocket) | G617E, G617V (AD-HIES) | G617R (Cancer) | T-LGLL (GOF) |
| Val637 | βD4 strand (pY+3 pocket) | V637M (AD-HIES) | - | Autosomal-dominant Hyper IgE Syndrome (LOF) |
This genetic volatility underscores the delicate evolutionary balance of wild-type STAT structural motifs in maintaining precise levels of cellular activity. The molecular basis for these opposing effects lies in how different substitutions at the same position alter the structural dynamics and binding properties of the SH2 domain [5].
Molecular Mechanisms of Opposing Functional Consequences
The structural context of mutational hotspots determines how different substitutions can produce opposing functional outcomes:
pY Pocket Mutations: Residues in the phosphate-binding pocket (e.g., Ser611, Ser614, Glu616, Gly617) directly impact phosphotyrosine binding affinity. Substitutions that disrupt key hydrogen bonds or charge interactions typically cause LOF, while mutations that enhance phosphate binding or stabilize active conformations can produce GOF [5].
Dimerization Interface Mutations: The SH2 domain mediates both phosphoreceptor binding and STAT dimerization. Mutations at the dimerization interface (e.g., αB, αB', BC* loop) can either impair or enhance dimer stability, with corresponding LOF or GOF effects [5].
Allosteric Regulatory Sites: Some hotspots reside in regions that allosterically regulate SH2 domain function, such as the hydrophobic core that maintains structural integrity. Disruptive substitutions cause LOF, while stabilizing mutations may produce GOF [5].
The BC loop region (residues 614-617 in STAT3) represents a particular hotspot cluster where subtle alterations dramatically impact function. For instance, STAT3 S614R (GOF) and S614G (LOF) mutations occur at identical positions but confer opposite effects on STAT3 transcriptional activity, with the arginine substitution potentially creating new salt bridges that enhance dimer stability [5].
Experimental Approaches for Investigating SH2 Domain Mutations
Structural Biology and Biophysical Techniques
X-ray Crystallography has been instrumental in characterizing STAT SH2 domain structures and mutations. The protocol typically involves:
- Recombinant Protein Expression: Clone STAT SH2 domain (residues ~575-700 for STAT3) into bacterial expression vectors with N-terminal His-tags for purification [66] [53]
- Site-Directed Mutagenesis: Introduce specific mutations using PCR-based methods (e.g., QuikChange) [66]
- Protein Purification: Purify proteins using nickel-affinity chromatography followed by size-exclusion chromatography [53]
- Crystallization: Screen crystallization conditions using commercial sparse matrix screens; STAT SH2 domains typically crystallize in PEG-containing conditions [66] [53]
- Data Collection and Structure Determination: Collect diffraction data at synchrotron sources and solve structures by molecular replacement [53]
Analytical Ultracentrifugation provides solution-state characterization of mutant effects on dimerization:
- Prepare mutant and wild-type STAT SH2 domains in buffer (e.g., 20mM HEPES, 150mM NaCl, pH 7.5)
- Conduct sedimentation equilibrium experiments at multiple speeds and protein concentrations
- Analyze data using non-linear least squares fitting to determine molecular weights and association constants [66]
Cellular Assays for Functional Characterization
FRET-Based Biosensors enable real-time monitoring of STAT activation and dimerization in live cells. The recently developed STATeLight biosensors represent a significant advancement [10]:
- Biosensor Design: C-terminally tag STAT with mNeonGreen (donor) and mScarlet-I (acceptor) fluorophores directly following the SH2 domain [10]
- Cell Line Development: Stably express biosensors in cytokine-responsive cells (e.g., HEK-Blue IL-2 cells for STAT5 studies) [10]
- FLIM-FRET Imaging: Measure fluorescence lifetime of the donor fluorophore; decreased lifetime indicates FRET and STAT activation [10]
- Stimulus Application: Treat cells with relevant cytokines (IL-6 for STAT3, IL-2 for STAT5) and monitor real-time activation kinetics [10]
- Mutant Analysis: Compare activation profiles of disease-associated mutants versus wild-type STAT [10]
Electrophoretic Mobility Shift Assays (EMSAs) assess DNA-binding capability:
- Prepare nuclear extracts from cytokine-stimulated cells expressing STAT mutants [4]
- Incubate extracts with ³²P-labeled double-stranded DNA probes containing GAS elements [4]
- Separate protein-DNA complexes from free probe using non-denaturing polyacrylamide gel electrophoresis [4]
- Visualize complexes by autoradiography or phosphorimaging [4]
Table 3: Research Reagent Solutions for STAT-SH2 Domain Investigations
| Reagent/Tool | Specifications | Experimental Applications | Key Features |
|---|---|---|---|
| STATeLight Biosensors | C-terminal mNeonGreen/mScarlet-I fusions | Real-time STAT activation monitoring via FLIM-FRET | High spatiotemporal resolution; compatible with live cells [10] |
| Recombinant SH2 Domains | 100-150 residue constructs with purification tags | Structural studies (crystallography, NMR); biophysical assays | Enables isolated domain characterization [66] [53] |
| Phosphopeptide Ligands | pY-containing peptides from receptor cytoplasmic domains | SH2 domain binding assays (SPR, ITC, fluorescence polarization) | Determines binding affinity and specificity [5] [8] |
| JAK/STAT Reporter Cells | Luciferase under GAS element control | High-throughput screening of STAT transcriptional activity | Quantifies functional output of signaling pathway [10] |
| Site-Directed Mutagenesis Kits | PCR-based systems (e.g., QuikChange) | Introduction of specific mutations into STAT constructs | Enables precise manipulation of hotspot residues [66] |
Visualization of STAT Activation and Mutational Impact
STAT Activation Pathway and Domain Organization
STAT Activation and SH2 Domain Function: This diagram illustrates the canonical STAT activation pathway, highlighting critical SH2 domain functions in receptor recruitment and phosphorylation-dependent dimerization.
Structural Consequences of SH2 Domain Mutations
Opposing Functional Consequences of SH2 Mutations: This workflow illustrates how mutations at identical positions in the SH2 domain can cause either loss-of-function or gain-of-function through distinct structural mechanisms.
Discussion: Therapeutic Implications and Future Directions
The delicate balance of STAT-SH2 domain function presents both challenges and opportunities for therapeutic development. The same structural features that make this domain a mutational hotspot also represent potential targets for precision medicine approaches. Several strategic considerations emerge:
Mutation-Specific Therapeutics: The opposing functional consequences of different substitutions at the same residue highlight the need for mutation-specific therapeutic strategies rather than gene-level interventions [5].
Allosteric Modulation: Targeting allosteric sites rather than the conserved pY pocket may offer better specificity for correcting pathological mutations without disrupting essential STAT functions [5] [8].
Context-Dependent Effects: Emerging evidence suggests that some SH2 domain mutations exhibit context-dependent effects, where the same mutation produces different outcomes in various cell types or developmental stages [5].
Future research directions should focus on:
- Comprehensive Structural Characterization: Expanding structural data for disease-associated STAT-SH2 mutants to understand precise mechanistic consequences [5]
- Dynamic Modeling: Employing molecular dynamics simulations to capture the flexibility and conformational landscapes of mutant SH2 domains [5] [8]
- Gene Editing Approaches: Developing CRISPR-based correction strategies that account for the delicate functional balance of hotspot residues [67]
- Chemical Biology Probes: Designing small molecules that specifically correct pathological mutations while preserving wild-type function [5] [8]
The STAT-SH2 domain exemplifies how precise evolutionary tuning of protein structural motifs can create functional landscapes where single amino acid changes produce dramatically opposed physiological outcomes. Understanding this delicate balance provides fundamental insights into molecular pathogenesis while revealing new opportunities for targeted therapeutic interventions in cancer, immunodeficiency, and autoimmune disorders.
The Signal Transducer and Activator of Transcription (STAT) family of proteins represents a critical signaling node that transmits information from extracellular cytokines and growth factors directly to the nucleus, regulating fundamental cellular processes including proliferation, differentiation, survival, and apoptosis. Comprising seven members (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6), these transcription factors share a conserved domain architecture featuring a central Src homology 2 (SH2) domain that is indispensable for their function [49] [33]. The SH2 domain facilitates critical protein-protein interactions by recognizing and binding to phosphotyrosine motifs, enabling STAT recruitment to activated cytokine receptors and subsequent tyrosine phosphorylation by Janus kinases (JAKs) [18] [68]. Following phosphorylation, STAT proteins form homo- or heterodimers through reciprocal SH2 domain-phosphotyrosine interactions, which then translocate to the nucleus to regulate gene expression [49] [33].
This whitepaper examines how alterations to the STAT-SH2 domain, whether through deliberate mutagenesis, natural mutations, or allosteric regulation, profoundly impact downstream signaling events, transcriptional programs, and ultimate cellular outcomes. Understanding these mechanistic relationships is crucial for both basic science and therapeutic development, particularly in cancer and immune disorders where STAT signaling is frequently dysregulated [68] [33]. We present integrated experimental evidence and computational analyses that delineate the molecular consequences of SH2 domain perturbations, providing a technical framework for researchers investigating STAT dimerization and its functional implications.
Canonical JAK-STAT Signaling and SH2 Domain Mechanics
The Standard Signaling Paradigm
The canonical JAK-STAT pathway represents a direct signaling cascade from cell surface receptors to nuclear gene transcription. In this established paradigm [49] [33]:
- Ligand-Receptor Engagement: Extracellular cytokines (e.g., interferons, interleukins) bind to their cognate transmembrane receptors, inducing receptor dimerization and activation of associated JAK kinases
- STAT Recruitment and Phosphorylation: Latent cytoplasmic STAT proteins are recruited to phosphorylated tyrosine residues on receptor cytoplasmic tails via their SH2 domains
- STAT Activation: JAKs phosphorylate a conserved tyrosine residue in the STAT C-terminal transactivation domain
- Dimerization and Nuclear Translocation: Phosphorylated STATs form homo- or heterodimers through reciprocal SH2 domain-phosphotyrosine interactions, then translocate to the nucleus
- Gene Transcription: STAT dimers bind specific DNA sequences (e.g., GAS elements for most STATs, ISRE elements for ISGF3 complex) to regulate target gene expression
Table 1: Core Components of Canonical JAK-STAT Signaling
| Component | Family Members | Key Functions |
|---|---|---|
| JAK Kinases | JAK1, JAK2, JAK3, TYK2 | Receptor-associated tyrosine kinases that phosphorylate STAT proteins |
| STAT Proteins | STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, STAT6 | SH2 domain-containing transcription factors that transduce signals to nucleus |
| Receptors | Type I/II cytokine receptors (IFN, IL, etc.) | Membrane receptors that dimerize upon ligand binding and activate JAKs |
Structural Basis of SH2 Domain Function
The STAT-SH2 domain is a highly conserved structural module of approximately 100 amino acids that facilitates phosphotyrosine-dependent protein-protein interactions [18] [49]. This domain contains critical structural features that enable its function:
- Phosphotyrosine (pY) Binding Pocket: A conserved arginine residue (R609 in STAT3) forms a crucial salt bridge with the phosphate group of phosphorylated tyrosine residues [69]
- Specificity-Determining Region: Residues that interact with amino acids C-terminal to the phosphotyrosine (pY+1, pY+2, pY+3) confer binding specificity [69]
- Dimerization Interface: Surface residues that mediate reciprocal interactions between STAT monomers during dimer formation [70] [71]
The SH2 domain enables two critical functions in STAT activation: first, it mediates temporary interactions with phosphorylated cytokine receptors during STAT recruitment and phosphorylation; second, it facilitates stable interactions between STAT monomers during dimer formation through reciprocal SH2-pY binding [49].
Figure 1: Canonical JAK-STAT Signaling Pathway. The diagram illustrates the sequential process from ligand-receptor engagement to STAT activation, dimerization via SH2 domain interactions, nuclear translocation, and target gene regulation.
SH2 Domain Mutations: Structural and Functional Consequences
Gain-of-Function Mutations in STAT-SH2 Domains
Specific mutations within the SH2 domain can result in constitutive or enhanced STAT signaling, fundamentally altering cellular responses to extracellular signals. Research has identified several critical residues where substitutions lead to gain-of-function (GOF) phenotypes:
STAT2 Y631F Mutation: A mutation in the conserved PYTK motif of the STAT2 SH2 domain (Y631F) demonstrates how single amino acid changes can dramatically alter signaling dynamics [70]. This mutation:
- Confers sustained tyrosine phosphorylation of STAT1 and STAT2 following IFN-α stimulation
- Prolongs the association of STAT1-STAT2 heterodimers
- Results in resistance to dephosphorylation by nuclear tyrosine phosphatase TcPTP
- Shifts cellular response from antiproliferative to apoptotic in tumor cells
STAT5B Y665F Mutation: Recent investigations of STAT5B mutations reveal similar GOF mechanisms [71]. The Y665F substitution:
- Enhances STAT5 phosphorylation, DNA binding, and transcriptional activity after cytokine activation
- Promotes accumulation of CD8+ effector and memory T cells and CD4+ regulatory T cells in mouse models
- Alters CD8+/CD4+ T cell ratios through enhanced signaling
- Represents a frequent mutation in T-cell large granular lymphoblastic leukemia (T-LGLL)
Table 2: Functional Consequences of SH2 Domain Mutations in STAT Proteins
| STAT Protein | Mutation | Structural Impact | Signaling Outcome | Cellular Phenotype |
|---|---|---|---|---|
| STAT2 | Y631F | Disrupts dephosphorylation resistance | Sustained signaling, prolonged ISGF3 activation | Apoptosis in tumor cells |
| STAT5B | Y665F | Stabilizes intramolecular interactions | Enhanced phosphorylation and DNA binding | Altered T-cell populations, GOF in leukemia |
| STAT5B | Y665H | Destabilizes C-terminal tail binding | Diminished phosphorylation, LOF | Reduced CD8+ effector and memory T cells |
Loss-of-Function Mutations and Allosteric Regulation
In contrast to GOF mutations, other SH2 domain alterations impair STAT function:
STAT5B Y665H Mutation: The same tyrosine residue can yield different functional outcomes depending on the substituting amino acid [71]. Unlike Y665F, the Y665H mutation:
- Introduces an imidazole group that destabilizes intramolecular interactions
- Displays loss-of-function (LOF) characteristics with diminished phosphorylation
- Results in reduced CD8+ effector and memory T cells and CD4+ regulatory T cells
- Demonstrates how subtle structural changes yield dramatically different functional outcomes
Allosteric Regulation of SH2 Domain: Beyond direct mutations, the SH2 domain function can be modulated allosterically through interactions with other STAT domains [69]. Molecular dynamics simulations reveal that:
- Perturbations in the coiled-coil domain (CCD) can induce conformational changes in the SH2 domain
- A rigid core comprising α-helices connects the CCD and SH2 domains via the linker domain
- Mutations like D170A in STAT3 CCD transmit long-range allosteric effects to the SH2 domain
- This allosteric regulation represents a potential therapeutic avenue for modulating STAT activity
Methodologies for Investigating SH2 Domain Function
Experimental Approaches for Characterizing SH2 Domain Mutations
Site-Directed Mutagenesis: Introduction of specific mutations into STAT genes enables functional characterization of SH2 domain residues [70]:
- Primer Design: Complementary primers containing desired mutations flanked by 10-15 base pairs of correct sequence on each side
- Template DNA: Flag-tagged STAT constructs in mammalian expression vectors (e.g., pcDNA3)
- Mutagenesis Kit: QuikChange XL Site-Directed Mutagenesis Kit or similar systems
- Verification: Sequencing the entire SH2 domain to confirm introduced mutations and exclude unintended changes
Cell Culture and Transfection Models: Cellular systems for evaluating STAT-SH2 function [70]:
- Parental Cell Line: Human fibrosarcoma 2fTGH cells
- STAT-Deficient Variants: U3A (STAT1−/−), U6A (STAT2−/−), and other isogenic derivatives
- Transfection Method: Metafectene reagent with STAT constructs (5 μg DNA)
- Selection: Stable clones maintained with G418 (500 μg/ml)
Functional Assays for STAT Signaling: Quantitative assessment of signaling outcomes [70]:
- Cell Proliferation: MTS assay in 96-well plates (1×10³ cells/well) with cytokine stimulation
- Apoptosis Measurement: Annexin V-FITC/propidium iodide staining with flow cytometry analysis
- Nuclear Translocation: Nuclear extract preparation and electrophoretic mobility shift assays (EMSAs)
- Gene Expression: Quantitative PCR for IFN-stimulated genes (ISGs)
Computational and Structural Biology Techniques
Molecular Dynamics Simulations: Advanced computational methods provide insights into SH2 domain dynamics [69]:
- System Setup: STAT3 structures (e.g., 1BG1 template) solvated in explicit water with physiological ion concentration
- Simulation Parameters: Energy minimization, equilibration, and production runs (100+ ns) using packages like AMBER or GROMACS
- Analysis Methods: Root-mean-square deviation (RMSD), principal component analysis (PCA), and dynamic cross-correlation matrices
- Variant Comparison: Wild-type STAT3 versus mutants (e.g., D170A) to identify conformational changes and allosteric pathways
In Silico Mutational Analysis: Predictive modeling of SH2 domain mutations [71]:
- Structure Prediction: AlphaFold3 for generating SH2 domain homodimer structures
- Energetic Calculations: COORDinator neural network for predicting stability effects of amino acid substitutions
- Pathogenicity Prediction: Tools including AlphaMissense, CADD, and REVEL for assessing mutation impact
- Conservation Analysis: Multiple sequence alignment across vertebrate species to identify evolutionarily constrained residues
Figure 2: Integrated Methodological Approaches for Investigating STAT-SH2 Domain Function. The workflow illustrates complementary experimental and computational techniques for characterizing SH2 domain mutations and their functional consequences.
Altered Transcriptional Programs and Cellular Outcomes
Prolonged Signaling and Apoptotic Responses
The sustained activation of STAT signaling resulting from certain SH2 domain mutations fundamentally alters transcriptional programs and cellular fate decisions. Research demonstrates that mutation-induced prolongation of STAT activation can shift cellular responses from growth inhibition to apoptosis [70]:
Temporal Control of Gene Expression:
- Wild-type STAT signaling: Transient phosphorylation (minutes to hours) with regulated termination
- Mutant STAT2 Y631F signaling: Sustained phosphorylation (>24 hours) with impaired dephosphorylation
- Enhanced duration of ISGF3 complex formation and nuclear retention
- Prolonged expression of interferon-stimulated genes (ISGs)
Dichotomous Cellular Outcomes:
- Antiproliferative response: Characteristic of wild-type STAT signaling in many tumor cell lines
- Apoptotic response: Induced by sustained signaling from SH2 domain mutants
- Correlation between signal duration and commitment to cell death pathways
- Therapeutic implications for manipulating STAT signaling dynamics in cancer
Immune Cell Differentiation and Function
SH2 domain mutations in STAT proteins significantly impact immune cell development, differentiation, and function through altered transcriptional programming [72] [71]:
T Helper Cell Polarization:
- STAT1 and STAT4: Critical for Th1 differentiation through response to IFN-γ and IL-12
- STAT6: Essential for Th2 differentiation via IL-4 signaling
- STAT3: Required for Th17 differentiation through induction of RORγt
- SH2 domain integrity necessary for proper lineage commitment
Cytotoxic T Cell and Treg Populations:
- STAT5B Y665F mutation increases CD8+ effector and memory T cell accumulation
- Altered CD8+/CD4+ T cell ratios through enhanced STAT5 signaling
- Expansion of CD4+ regulatory T cells with GOF STAT5 mutations
- Implications for autoimmune disease and cancer immunotherapy
Table 3: Research Reagent Solutions for STAT-SH2 Domain Investigations
| Reagent Category | Specific Examples | Research Applications |
|---|---|---|
| Cell Line Models | 2fTGH and variants (U3A/STAT1−/−, U6A/STAT2−/−) | Isogenic systems for STAT functional studies |
| Mutagenesis Kits | QuikChange XL Site-Directed Mutagenesis Kit | Introduction of specific SH2 domain mutations |
| STAT Constructs | Flag-tagged STAT2 in pcDNA3 | Expression vectors for reconstitution experiments |
| Cytokines | Recombinant human IFN-α-2a, IFN-β, IFN-γ | STAT pathway activation and stimulation |
| Apoptosis Assays | Annexin V-FITC/propidium iodide staining | Quantification of cell death responses |
| Computational Tools | AlphaFold3, COORDinator, Molecular Dynamics | Predicting structural impacts of SH2 mutations |
Therapeutic Implications and Future Directions
Targeting SH2 Domains in Disease
The critical role of STAT-SH2 domains in dimerization and signaling makes them attractive therapeutic targets, particularly in cancer and autoimmune diseases [68] [33]:
Oncogenic STAT Signaling:
- Constitutive STAT activation observed in diverse hematologic malignancies and solid tumors
- SH2 domain mutations identified in T-cell leukemias (STAT5B N642H, Y665F)
- Direct SH2 domain inhibitors prevent STAT dimerization and nuclear translocation
- Challenges with specificity and pharmacologic efficacy in clinical development
Allosteric Modulation Strategies:
- Targeting coiled-coil or other domains to indirectly modulate SH2 function
- Small molecules (MM-206, K116) and polypeptides (MS3-6) that bind CCD and affect SH2 conformation
- Potential for enhanced specificity compared to direct SH2 inhibitors
- At least one CCD-targeting drug candidate in Phase I clinical trials [69]
Research Applications and Technical Considerations
For researchers investigating STAT-SH2 domain function, several technical considerations emerge from current literature:
Experimental Design:
- Employ multiple complementary approaches (cellular, biochemical, computational)
- Include both gain-of-function and loss-of-function mutants for comparative analysis
- Consider temporal aspects of signaling in addition to magnitude of activation
- Evaluate both nuclear/transcriptional and non-canonical STAT functions
Interpretation Challenges:
- Context-dependent effects of STAT mutations across cell types and stimuli
- Potential opposing functions of phosphorylated versus unphosphorylated STAT pools
- Complex interplay between different STAT family members and negative regulators
- Species-specific differences in STAT signaling and immune function
The STAT-SH2 domain represents a critical structural and functional module that governs dimerization, nuclear translocation, and transcriptional activity of STAT proteins. Mutations or targeted perturbations within this domain can dramatically alter downstream signaling events, transcriptional programs, and ultimate cellular outcomes. The research findings synthesized in this technical review demonstrate that single amino acid changes in the SH2 domain can convert transient signaling responses into sustained activation, shift cellular fate decisions from proliferation to apoptosis, and reshape immune cell populations through altered differentiation pathways.
Future research directions should focus on elucidating the allosteric networks connecting the SH2 domain to other STAT functional regions, developing more specific therapeutic agents that target pathological STAT signaling while preserving physiological functions, and exploring the context-dependent outcomes of STAT activation across different tissue and disease environments. The integrated methodological approaches combining computational predictions with experimental validation provide a powerful framework for advancing our understanding of STAT-SH2 domain biology and its translational applications.
The canonical activation of Signal Transducers and Activators of Transcription (STATs) through tyrosine phosphorylation and SH2 domain-mediated dimerization is a well-established paradigm. However, emerging research reveals a complex landscape of STAT regulation that extends beyond this classical mechanism. Unphosphorylated STATs (U-STATs) form dimers through alternative domains and exert distinct biological functions, contributing to tumorigenesis, immune regulation, and cellular homeostasis. This whitepaper synthesizes current understanding of U-STAT dimer formation, their molecular regulation, and pathological significance, with particular emphasis on implications for STAT-SH2 domain research and therapeutic targeting. The dysregulation of U-STAT dimers represents a previously underappreciated layer of complexity in cellular signaling with profound implications for drug discovery.
The JAK-STAT signaling pathway serves as a critical communication node for numerous cytokines and growth factors, regulating essential processes including hematopoiesis, immunity, and cell proliferation [33]. Canonical STAT activation involves tyrosine phosphorylation by Janus kinases (JAKs) at specific C-terminal residues, triggering SH2 domain-mediated reciprocal dimerization, nuclear translocation, and DNA binding to regulate target gene expression [33] [32].
Historically, STAT research has focused heavily on phosphorylated, dimeric forms, with the SH2 domain recognized as essential for this activation mechanism. The SH2 domain, approximately 100 amino acids in length, contains a highly conserved arginine residue that forms a salt bridge with phosphotyrosine motifs, enabling specific protein-protein interactions [16] [15] [8]. In phosphorylated STAT dimers, the SH2 domain of one monomer binds the phosphorylated tyrosine of its partner, creating a stable dimeric complex [32].
However, compelling evidence now demonstrates that STAT proteins form dimers and exert biological functions independently of tyrosine phosphorylation. These U-STAT dimers utilize distinct molecular interfaces, primarily through N-terminal domains (NTDs), and regulate gene expression programs that often differ from their phosphorylated counterparts [73]. The dysregulation of U-STAT dimers contributes significantly to pathological states, particularly cancer, presenting new challenges and opportunities for therapeutic intervention.
Molecular Architecture of STAT Proteins and Dimerization Interfaces
STAT proteins share a conserved domain structure that facilitates their diverse functions. Understanding this architecture is fundamental to appreciating the distinct mechanisms of phosphorylated versus unphosphorylated dimer formation.
STAT Domain Organization
All mammalian STAT proteins contain six conserved functional domains:
- N-terminal domain (NTD): Facilitates tetramerization, nuclear import, and protein-protein interactions
- Coiled-coil domain: Mediates interactions with regulatory proteins and nuclear export
- DNA-binding domain: Recognizes specific DNA sequences
- Linker domain: Maintains conformational stability
- SH2 domain: Binds phosphorylated tyrosine residues
- Transactivation domain (TAD): Regulates transcriptional activity [73] [74]
Table 1: STAT Protein Domains and Their Functions
| Domain | Primary Functions | Role in Phosphorylated Dimers | Role in Unphosphorylated Dimers |
|---|---|---|---|
| N-terminal domain (NTD) | Tetramerization, nuclear import | Accessory role | Primary dimerization interface |
| SH2 domain | Phosphotyrosine recognition | Essential for dimer stability | Limited involvement |
| DNA-binding domain | Target gene recognition | Required for function | Required for function |
| Transactivation domain (TAD) | Transcriptional activation | Phosphorylation-dependent | Phosphorylation-independent |
Structural Basis for Alternative Dimerization
The SH2 domain engages in canonical STAT dimerization through a highly specific interaction where the SH2 domain of one monomer binds the phosphorylated tyrosine (pY) of its partner, forming a contiguous C-shaped clamp around DNA [32]. This configuration is stabilized by reciprocal interactions between the SH2 domain and C-terminal phosphotyrosine segment.
In contrast, U-STAT dimers primarily utilize N-terminal domains for dimerization. Crystallographic studies reveal that STAT NTDs fold independently and contain conserved residues critical for oligomerization. The invariant tryptophan residue at position 37 (W37) is essential for ND dimerization, though other residues including Q36, T40, E66, S12, L15, DR19, F77, and L78 contribute to the interaction interface [73]. Unlike phosphorylated dimers that form stable complexes through SH2-pY interactions, U-STAT dimers demonstrate more dynamic association properties, potentially enabling more rapid response to cellular signals.
Formation and Regulation of Unphosphorylated STAT Dimers
Molecular Mechanisms of U-STAT Dimerization
U-STAT dimerization occurs through domain interactions distinct from the canonical SH2-phosphotyrosine pairing. The primary mechanism involves N-terminal domain interactions that enable STAT proteins to form dimers independently of activation loop phosphorylation.
Research indicates that STAT NTDs mediate tetramer formation when two STAT dimers bind adjacent DNA sites [73]. This tetramerization function appears later in evolution, present in Drosophila, zebrafish, and mammalian STATs but absent in simpler organisms like Dictyostelium and C. elegans [73]. The accretion of NTDs during evolution may have provided STAT proteins with enhanced functional flexibility in DNA binding and transcriptional regulation.
The functional significance of ND-mediated oligomerization is demonstrated by several key findings:
- STAT1 N-terminal domain mutations (F77) impair oligomerization and dephosphorylation, reducing antiviral protection conferred by IFNα [73]
- STAT4 with W37A substitution prevents IFNα-induced tyrosine phosphorylation, blocking both dimer and tetramer formation [73]
- STAT3 N-terminal domains are essential for driving cancer cell proliferation and survival [73]
Distinct Gene Regulatory Programs
U-STAT dimers regulate transcriptional programs that often differ substantially from those activated by phosphorylated STATs. This functional divergence stems from several factors:
DNA Binding Specificity: While phosphorylated STAT dimers bind canonical GAS (gamma-activated sequence) elements, U-STAT dimers demonstrate altered DNA binding affinity and specificity. For instance, STAT3 tetramerization via N-terminal domains is required for α2-macroglobulin gene promoter activation but dispensable for SOCS3 activation [73].
Temporal Expression Patterns: U-STAT levels often increase later than phosphorylated STATs following pathway activation. For example, phosphorylated STAT3 stimulates its own transcription, causing increased U-STAT3 levels that subsequently contribute to tumorigenesis through distinct mechanisms [73].
Differential Partner Interactions: U-STAT dimers engage different protein interaction partners compared to phosphorylated STATs, leading to alternative transcriptional complexes and target gene specificity.
Table 2: Comparative Functions of Phosphorylated STATs vs. U-STATs
| Feature | Phosphorylated STATs | Unphosphorylated STATs |
|---|---|---|
| Dimerization Interface | SH2 domain-phosphotyrosine | N-terminal domain |
| Nuclear Translocation | Signal-dependent | Constitutive |
| DNA Binding Specificity | High-affinity for GAS sites | Broader, weaker specificity |
| Transcriptional Output | Acute, transient response | Sustained, alternative programs |
| Primary Biological Role | Cytokine-responsive signaling | Homeostatic regulation, chronic adaptation |
| Pathological Significance | Acute inflammation, proliferation | Tumor maintenance, therapy resistance |
Pathological Significance of U-STAT Dimers
Oncogenic Roles in Cancer
U-STAT dimers play particularly significant roles in oncogenesis, often contributing to phenotypes distinct from those driven by phosphorylated STATs:
STAT3 in Carcinogenesis: Constitutively activated STAT3 increases U-STAT3 levels through transcriptional auto-regulation. This U-STAT3 pool contributes to tumorigenesis through mechanisms different from phosphorylated STAT3, including regulation of distinct target genes that enhance malignant potential [73]. In clear cell renal cell carcinoma, non-canonical phosphorylation at Ser727 enhances STAT3's DNA-binding affinity to promoters of invasion-related genes like VEGF and CXCL1 [74].
STAT5 in Leukemogenesis: The causative role of STAT5 N-terminal domain in leukemogenesis has been experimentally demonstrated [73]. STAT5 tetramerization, mediated through N-terminal domains, is essential for IL-2-induced regulation of the IL-2 response element in the human IL-2Ra gene [73].
Therapeutic Resistance: U-STAT dimers contribute to resistance mechanisms against targeted therapies. In chronic myeloid leukemia (CML), leukemia stem cells persist despite tyrosine kinase inhibitor treatment through survival mechanisms that are not entirely reliant on BCR-ABL activation [75]. These persistent cells likely utilize alternative signaling pathways, potentially involving U-STAT dimers.
Role in Immune Regulation
U-STAT dimers participate in immune system regulation through several mechanisms:
Interferon Signaling Modulation: Sendai virus C protein exemplifies how pathogens exploit U-STAT dimerization to evade host immunity. The C-terminal region of C protein (Y3) binds directly to the N-terminal domain of STAT1 (STAT1ND), inhibiting IFN-α/β-induced STAT1:STAT2 pathway and IFN-γ-induced STAT1 pathway [76]. Interestingly, C protein also binds STAT3ND, though with weaker affinity, stimulating IFN-γ-induced STAT3 phosphorylation and creating persistent STAT3 pathway activation [76].
Cytokine Response Fine-Tuning: The requirement for STAT tetramerization via NTD contributes to selective activation of certain genes. For example, STAT5 tetramerization is necessary for IL-2Ra expression but dispensable for β-casein expression [73]. This gene-specific requirement enables precise immune response modulation.
Experimental Approaches for Studying U-STAT Dimers
Methodologies for Detection and Characterization
Yeast Two-Hybrid Analysis: This technique identifies protein-protein interactions through transcriptional activation of reporter genes. Studies using STAT N-terminal domains as bait have revealed novel interaction partners, including the Sendai virus C protein binding to STAT1ND and STAT3ND [76].
Protocol: Yeast Two-Hybrid Screening for STAT ND Interactions
- Clone STAT N-terminal domain into bait vector (DNA-binding domain fusion)
- Transform bait construct into appropriate yeast strain
- Mate with prey library (activation domain fusion)
- Select for interactions on appropriate dropout media
- Confirm positive interactions through β-galactosidase assay
- Sequence prey plasmids to identify interacting partners
Size Exclusion Chromatography with Multi-Angle Light Scattering (SEC-MALS): This technique determines molecular mass and oligomeric states under native conditions.
Protocol: SEC-MALS Analysis of STAT Oligomerization
- Express and purify recombinant STAT N-terminal domains
- Pre-equilibrate SEC column with appropriate buffer
- Inject protein sample at concentrations ranging 8-160 μM
- Monitor elution with UV detector, light scattering, and refractive index
- Calculate molecular mass using ASTRA or equivalent software
- Compare observed mass with theoretical values to determine stoichiometry
Native Mass Spectrometry: This method directly analyzes intact protein complexes under non-denaturing conditions, providing precise stoichiometry information.
Protocol: Native MS for Complex Stoichiometry
- Desalt protein samples into volatile ammonium acetate buffer
- Introduce sample via nano-electrospray ionization
- Optimize instrument parameters to preserve non-covalent interactions
- Acquire mass spectra under native conditions
- Deconvolute spectra to determine complex molecular weights
- Calculate binding stoichiometry from mass differences
Functional Assays
Reporter Gene Assays: These experiments measure transcriptional activity of U-STAT dimers by linking STAT-responsive promoters to easily quantifiable reporters.
Protocol: Reporter Assay for U-STAT Activity
- Clone STAT-responsive promoter elements upstream of luciferase gene
- Co-transfect reporter construct with U-STAT expression vectors
- Include empty vector and phosphorylation-deficient mutants as controls
- Harvest cells 24-48 hours post-transfection
- Measure luciferase activity using luminometer
- Normalize to co-transfected control reporter (e.g., Renilla luciferase)
Cellular Localization Studies: These investigations track subcellular distribution of U-STAT dimers using imaging techniques.
Protocol: Confocal Microscopy for STAT Localization
- Express fluorescently tagged U-STAT constructs in appropriate cell lines
- Stimulate with relevant cytokines or inhibitors as experimental conditions require
- Fix cells at designated time points
- Counterstain nuclei with DAPI or Hoechst dyes
- Image using confocal microscope with appropriate filter sets
- Quantify nuclear/cytoplasmic distribution using image analysis software
Research Reagent Solutions
Table 3: Essential Research Reagents for U-STAT Dimer Investigation
| Reagent/Category | Specific Examples | Research Application | Key Considerations |
|---|---|---|---|
| Expression Vectors | pEYFP-STAT3, pGFP-STAT1 | Fluorescent tagging for localization | Select full-length vs. domain-specific constructs |
| Antibodies | Anti-STAT1 (phospho-Y701), Total STAT3 | Differentiation between phosphorylated and U-STAT forms | Validate specificity for Western blot, IF, IHC |
| Cell Lines | STAT-deficient U3A, MEFs from knockout mice | Background reduction for clean readouts | Verify genotype and functional STAT deficiency |
| Recombinant Proteins | Purified STAT N-terminal domains | Biophysical characterization | Ensure proper folding and post-translational modifications |
| Chemical Inhibitors | AG490 (JAK2 inhibitor), Stattic (STAT3 inhibitor) | Dissect phosphorylation-dependent and independent functions | Optimize concentration to avoid off-target effects |
Therapeutic Implications and Targeting Strategies
Challenges in Targeting U-STAT Dimers
The development of therapeutics targeting U-STAT dimers presents unique challenges:
Domain Specificity: STAT N-terminal domains do not share significant homology with other proteins, reducing potential off-target effects but complicating drug design due to limited structural information [73].
Functional Redundancy: Different STAT family members can exhibit overlapping functions, potentially necessitating multi-target approaches for effective inhibition.
Differential Roles in Physiology vs. Pathology: U-STAT dimers likely serve important physiological functions, requiring strategies that selectively inhibit pathological without disrupting normal functions.
Emerging Targeting Approaches
Peptide-Based Inhibitors: Lipopeptides mimicking N-terminal domain interfaces can disrupt U-STAT dimerization. Rational design approaches utilize structural information to develop stabilized peptides with enhanced cellular permeability and binding affinity [73].
Small Molecule Inhibitors: High-throughput screening approaches identify compounds that specifically disrupt ND-mediated STAT interactions. The unique structural features of STAT NDs offer opportunities for selective targeting compared to the more conserved SH2 domains.
Combination Therapies: Simultaneously targeting both phosphorylated and unphosphorylated STAT pools may provide enhanced efficacy, particularly in resistant cancers. For example, in chronic myeloid leukemia, targeting downstream pathways essential for leukemia stem cell survival may overcome limitations of tyrosine kinase inhibitors [75].
Visualizing U-STAT Dynamics and Experimental Approaches
The following diagrams illustrate key concepts and methodologies in U-STAT dimer research.
Diagram 1: STAT Dimerization Mechanisms
Diagram 2: Experimental Workflow for U-STAT Dimer Characterization
The investigation of unphosphorylated STAT dimers represents a paradigm shift in understanding STAT biology and its pathological dysregulation. While the SH2 domain remains central to canonical STAT activation, alternative dimerization mechanisms through N-terminal domains significantly expand the functional repertoire of STAT proteins. The dysregulation of U-STAT dimers contributes to disease states, particularly cancer, through distinct transcriptional programs that often complement and reinforce phenotypes driven by phosphorylated STATs.
Future research directions should include comprehensive structural characterization of U-STAT dimers, identification of specific gene targets, and development of selective inhibitors that disrupt pathological U-STAT functions without compromising physiological signaling. Integration of U-STAT dimer investigation into STAT research frameworks will provide a more complete understanding of cellular signaling networks and open new avenues for therapeutic intervention in cancer and other diseases characterized by STAT pathway dysregulation.
Therapeutic Targeting and Comparative Analysis of STAT Family Dimers
The Src homology 2 (SH2) domain is a protein module of approximately 100 amino acids that specifically recognizes and binds to phosphorylated tyrosine (pY) motifs, serving as a critical mediator in intracellular signaling networks [8]. These domains are found in approximately 110 human proteins, including enzymes, adaptors, transcription factors, and regulators of cytoskeletal dynamics [8]. The ability of SH2 domains to facilitate precise protein-protein interactions through pY recognition places them at the heart of numerous signaling pathways that control essential cellular processes such as proliferation, differentiation, survival, and immune responses [8] [77]. Among the most structurally and functionally significant SH2 domain-containing proteins are the Signal Transducers and Activators of Transcription (STATs), particularly STAT3 and STAT5, whose dimerization and transcriptional activity depend critically on their SH2 domains [4] [6].
The therapeutic targeting of SH2 domains represents a promising strategy for modulating pathological signaling cascades, especially in cancer and immune disorders. Unlike kinase domains, which share considerable structural homology, SH2 domains offer greater potential for selective inhibition due to their more diverse pY-binding pockets [8] [78]. This review examines the challenges and opportunities in targeting SH2 domains, with a specific focus on the STAT-SH2 domain in STAT dimerization. We synthesize recent structural insights, profiling emerging inhibitory modalities, and detailing experimental approaches for evaluating SH2 domain function and inhibition.
Structural Basis of SH2 Domain Function
Conserved Architecture and Phosphotyrosine Recognition
All SH2 domains share a conserved structural fold characterized by a central three-stranded antiparallel β-sheet flanked by two α-helices, forming a compact "sandwich" structure [8]. Despite sometimes sharing as little as 15% sequence identity across family members, this core three-dimensional architecture remains remarkably conserved, underscoring its fundamental role in pY recognition [8]. The N-terminal region of the SH2 domain contains a deep pocket that binds the phosphate moiety of phosphorylated tyrosine residues. This pocket harbors an absolutely conserved arginine residue at position βB5 (part of the FLVR motif) that forms a critical salt bridge with the phosphate group [8].
Table 1: Key Structural Elements of SH2 Domains
| Structural Element | Location | Functional Role |
|---|---|---|
| Central β-sheet | Core domain | Provides structural scaffold |
| αA and αB helices | Flanking β-sheet | Contribute to structural stability |
| pY-binding pocket | N-terminal region (βB strand) | Binds phosphate moiety via conserved arginine |
| Specificity pocket | C-terminal region | Determines sequence specificity C-terminal to pY |
| EF and BG loops | Variable regions | Influence phosphopeptide binding affinity |
Beyond the conserved pY-binding site, the C-terminal region of SH2 domains contains structural elements that determine sequence specificity for residues C-terminal to the phosphotyrosine. The length and conformation of intervening loops, particularly the EF loop (joining β-strands E and F) and BG loop (joining α-helix B and β-strand G), vary among SH2 domains and contribute to ligand specificity [8]. This structural diversity enables different SH2 domains to recognize distinct pY-containing motifs, allowing for precise signaling specificity within complex cellular environments.
Non-Canonical Functions: Lipid Binding and Phase Separation
Emerging research reveals that SH2 domains possess functions beyond pY recognition. Nearly 75% of SH2 domains interact with membrane lipids, particularly phosphoinositides such as PIP₂ and PIP₃ [8]. These interactions often occur through cationic regions near the pY-binding pocket that are flanked by aromatic or hydrophobic residues, forming distinct lipid-binding sites [8]. For example, the PIP₃ binding activity of the TNS2 SH2 domain regulates phosphorylation of insulin receptor substrate-1 (IRS-1) in insulin signaling, while lipid binding by SYK, ZAP70, and VAV2 SH2 domains modulates their membrane recruitment and enzymatic activities [8].
SH2 domain-containing proteins have also been implicated in the formation of biomolecular condensates through liquid-liquid phase separation (LLPS) [8]. The multivalent interactions facilitated by SH2 domains and other modular domains drive the assembly of these membrane-less organelles, which enhance signaling specificity and efficiency. In T-cell receptor signaling, interactions among GRB2, Gads, and the LAT adapter protein contribute to LLPS formation, creating concentrated signaling hubs that optimize signal transduction [8]. Similarly, in kidney podocytes, phase separation of the NCK adapter protein increases membrane dwell time of N-WASP–Arp2/3 complexes, promoting actin polymerization [8]. These non-canonical functions expand the potential therapeutic applications of SH2 domain targeting beyond simple disruption of protein-protein interactions.
The STAT-SH2 Domain in STAT Dimerization
Structural Mechanism of STAT Activation and Dimerization
STAT proteins are latent cytoplasmic transcription factors that become activated by cytokines, growth factors, and hormones. Among the seven STAT family members, STAT3 and STAT5 are particularly important in oncogenesis and immune regulation [4] [79]. The canonical activation pathway begins when extracellular ligands bind to their cognate receptors, triggering receptor dimerization and activation of associated Janus kinases (JAKs) [10] [6]. The JAKs phosphorylate tyrosine residues on the receptor intracellular domains, creating docking sites for STAT proteins via their SH2 domains [6].
Once recruited, STATs undergo JAK-mediated phosphorylation at a conserved tyrosine residue (Y705 in STAT3, Y694/Y699 in STAT5A/5B) within their SH2 domains [10] [6]. This phosphorylation initiates a dramatic conformational change: STAT monomers transition from "antiparallel" dimers in the inactive state to "parallel" dimers in the active state [10]. The phosphorylated tyrosine of one STAT monomer inserts into the SH2 domain of its partner, forming stable reciprocal SH2-pY interactions that stabilize the active dimer [6]. These phosphorylated STAT dimers then translocate to the nucleus, where they bind gamma-activated sequence (GAS) elements in target gene promoters and initiate transcription [4] [10].
Figure 1: STAT Activation Pathway and SH2 Domain-Mediated Dimerization. STAT activation begins with cytokine binding, leading to JAK-mediated phosphorylation and reciprocal SH2-pY dimer formation.
Role of the SH2 Domain in STAT Regulation
The SH2 domain serves as the central regulatory module controlling STAT dimerization and activation. Beyond facilitating the reciprocal interactions in active dimers, the SH2 domain mediates nuclear translocation of STAT dimers through interactions with importin proteins [4]. Recent research has also revealed that unphosphorylated STATs (U-STATs) can form dimers and regulate gene expression through SH2 domain-dependent mechanisms that differ from canonical phosphorylation-dependent signaling [4].
For STAT3, the Cys367-Cys542 disulfide bridge within the SH2 domain is essential for the DNA-binding activity of unphosphorylated STAT3 (U-STAT3) [4]. Mutation of these cysteine residues completely abolishes U-STAT3 DNA-binding capacity, indicating that this disulfide bridge induces structural changes that alter the overall conformation of dimeric U-STAT3 species [4]. U-STAT3 can bind to both GAS elements and AT-rich DNA sequences, functioning as both a transcriptional activator and a chromatin organizer [4].
Emerging Strategies for Targeting SH2 Domains
Small Molecule Inhibitors
The development of small molecule inhibitors targeting SH2 domains has gained significant momentum, with several promising compounds emerging in recent years. These inhibitors typically function by occupying the pY-binding pocket, thereby competitively blocking native protein-protein interactions. For the STAT3 SH2 domain, compounds 323-1 and 323-2 (delavatine A stereoisomers) have demonstrated potent inhibition of both phosphorylated and non-phosphorylated STAT3 dimerization by directly binding to the SH2 domain [6]. Computational docking predicts that these compounds bind to three subpockets of the STAT3 SH2 domain, exhibiting stronger inhibition than the commercial STAT3 inhibitor S3I-201 in co-immunoprecipitation assays [6].
In fluorescence polarization assays, compounds 323-1 and 323-2 competitively abrogated the interaction between STAT3 and the SH2-binding peptide GpYLPQTV, confirming their mechanism of action [6]. These compounds also demonstrated cellular efficacy, reducing IL-6-stimulated phosphorylation of STAT3 at Tyr705 in LNCaP cells and downregulating STAT3 target genes MCL1 and cyclin D1 [6]. Compared to S3I-201, the 323 compounds showed superior selectivity for STAT3 over STAT1 phosphorylation, highlighting their potential as lead compounds for cancers with hyperactivated STAT3 [6].
Peptide-Based Inhibitors and Monobodies
Peptide-based approaches and engineered protein scaffolds offer alternative strategies for targeting SH2 domains with high specificity. For the SHP2 phosphatase, which contains two SH2 domains (N-SH2 and C-SH2), researchers have developed a C-SH2 inhibitor peptide (CSIP) incorporating a non-hydrolysable pTyr mimetic, l-O-malonyltyrosine (l-OMT) [80]. This peptide selectively binds to the C-SH2 domain of SHP2 and blocks its protein-protein interactions [80]. Interestingly, incorporation of the widely used pTyr mimetic phosphonodifluoromethyl phenylalanine (F2Pmp) abolished binding, challenging existing notions about general SH2 domain binders [80]. The CSIP exhibits excellent stability, selectivity, cell permeability, and non-cytotoxicity, enriching the toolbox of SHP2 inhibitors with different modes of action [80].
Monobodies – synthetic binding proteins based on the fibronectin type III domain – represent another innovative approach. Researchers have engineered a monobody termed Mb(SHP2PTP_13) (Mb13) that specifically binds to the protein tyrosine phosphatase (PTP) domain of SHP2 [81]. Extensive molecular dynamics simulations revealed that Mb13 binds more stably to SHP2-PTP compared to SHP1-PTP, with the SHP2 complex exhibiting conformational stability and reduced flexibility [81]. Detailed analysis identified key residues within SHP2-PTP that form robust interactions with Mb13, driving the selective binding mechanism [81].
BTK SH2 Domain Inhibition: A Case Study in Selectivity
Recent breakthroughs in targeting the BTK SH2 domain illustrate the potential for achieving exceptional selectivity. Recludix Pharma has developed the first-known inhibitors of the BTK SH2 domain, demonstrating best-in-class selectivity that exceeds even the most selective kinase domain inhibitors [78]. Unlike traditional BTK kinase inhibitors (such as ibrutinib), which often exhibit off-target effects including platelet dysfunction due to TEC kinase inhibition, BTK SH2 inhibitors (BTK SH2i) avoid these complications through their unique mechanism [78].
The lead BTK SH2i compound exhibits remarkable biochemical potency (BTK Kd = 0.055 nM) with minimal cytotoxicity (>10,000 nM EC50 in Jurkat cells) and exceptional SH2ome selectivity (>8000-fold over off-target SH2 domains) [78]. In cellular assays, BTK SH2i robustly inhibits SH2-dependent pERK signaling and suppresses downstream CD69 expression in B cells and TMD8 lymphoma cells [78]. Pharmacokinetic studies in dogs demonstrated sustained intracellular concentrations and prolonged BTK target engagement over 48 hours following intravenous dosing [78]. In a mouse model of ovalbumin-induced chronic spontaneous urticaria (CSU), a single prophylactic dose of BTK SH2i led to significant, dose-dependent reduction in skin inflammation, outperforming remibrutinib and ibrutinib in suppressing vascular leakiness and inflammatory cell infiltration [78].
Table 2: Comparison of SH2 Domain-Targeting Therapeutic Approaches
| Therapeutic Approach | Molecular Target | Key Characteristics | Development Status |
|---|---|---|---|
| Small Molecules (323-1, 323-2) | STAT3 SH2 domain | Inhibit phosphorylated and non-phosphorylated STAT3 dimerization; target multiple subpockets | Preclinical |
| Peptide Inhibitors (CSIP) | SHP2 C-SH2 domain | Selective C-SH2 binding with non-hydrolysable pTyr mimetic; cell permeable and non-cytotoxic | Preclinical |
| Monobodies (Mb13) | SHP2 PTP domain | High specificity through conformational stabilization; distinct from SH2 direct targeting | Preclinical |
| BTK SH2 Inhibitors (Recludix) | BTK SH2 domain | Exceptional selectivity (>8000-fold); avoids TEC kinase inhibition; durable pathway inhibition | Preclinical |
Experimental Approaches for Studying SH2 Domain Function and Inhibition
Biosensor Development for Real-Time Monitoring
Innovative biosensor technologies have emerged to enable real-time monitoring of STAT activation and dimerization in live cells. Researchers have developed STATeLights – genetically encoded biosensors based on FRET (Förster resonance energy transfer) that allow direct and continuous detection of STAT activity with high spatiotemporal resolution [10]. These biosensors utilize fluorescence lifetime imaging microscopy (FLIM)-FRET, where FRET efficiency is inversely correlated to the fluorescence lifetime of the donor fluorophore, providing advantages over conventional ratiometric FRET approaches due to limited dependency on fluorophore expression level and photobleaching [10].
The STATeLight biosensor for STAT5A was engineered by tagging STAT5A monomers with mNeonGreen (donor) and mScarlet-I (acceptor) fluorescent proteins at optimal positions to detect cytokine-mediated conformational changes from antiparallel to parallel dimers [10]. Through comprehensive screening of various fusion constructs, the most effective design involved C-terminal fusion of fluorescent proteins to truncated STAT5A containing the core fragment plus the C-terminus [10]. This configuration demonstrated up to 12% FRET efficiency upon IL-2 stimulation, consistent with the predicted close distance between SH2 domains in the parallel conformation [10]. STATeLights enable specific observation of STAT activation by directly monitoring conformational rearrangement of STAT dimers, avoiding potential adverse signals from inactive phosphorylated monomers or truncated STAT variants [10].
Figure 2: STAT Biosensor Operation. The STATeLight biosensor detects dimerization through FRET efficiency changes between fluorescent proteins fused to STAT monomers.
Molecular Dynamics and Computational Approaches
Computational methods, particularly molecular dynamics (MD) simulations, have become indispensable tools for understanding SH2 domain dynamics and inhibitor mechanisms. In studies of the SHP2 phosphatase, extensive MD simulations of the Mb13-SHP2-PTP and Mb13-SHP1-PTP systems, combined with cluster analysis, principal component analysis, free energy landscape evaluation, and binding free energy calculations, revealed that Mb13 binds more stably to SHP2-PTP compared to SHP1-PTP [81]. The SHP2 complex exhibited reduced flexibility and conformational stability, indicating stronger interactions [81]. These computational approaches identified specific residue interactions that drive selective binding, providing atomic-level insights into the inhibition mechanism [81].
Similarly, computational docking studies have been essential for understanding how small molecules like compounds 323-1 and 323-2 interact with the STAT3 SH2 domain [6]. These predictions guide rational drug design by identifying key binding subpockets and interaction residues, enabling optimization of inhibitor potency and selectivity.
Disease Modeling with Genetic Mutations
The functional significance of SH2 domains is further highlighted by disease-associated mutations that disrupt normal regulation. For STAT5B, mutations at tyrosine 665 (Y665) within the SH2 domain have profound physiological consequences [79]. The Y665H mutation functions as a loss-of-function mutation, impairing enhancer establishment and alveolar differentiation in mammary gland development, resulting in lactation failure [79]. Conversely, the Y665F mutation acts as a gain-of-function mutation, elevating enhancer formation and accelerating mammary development during pregnancy [79].
These mutations have been modeled in mice using CRISPR/Cas9 and base editing technologies, providing insights into how single amino acid alterations in SH2 domains modulate genetic programs in hormonally regulated signaling pathways [79]. Transcriptomic and epigenomic analyses of these mutants revealed their opposing effects on enhancer landscapes and gene expression programs, underscoring the critical role of the SH2 domain in regulating STAT5B transcriptional output [79].
The Scientist's Toolkit: Essential Research Reagents and Methodologies
Table 3: Research Reagent Solutions for SH2 Domain Studies
| Research Tool | Specific Example | Application/Function | Experimental Context |
|---|---|---|---|
| FRET-Based Biosensors | STATeLight5A | Real-time monitoring of STAT5 activation via FLIM-FRET | Live-cell imaging of STAT dimerization [10] |
| Fluorescent Protein Pairs | mNeonGreen/mScarlet-I | Donor/acceptor FRET pair with optimal spectral properties | Biosensor engineering for STAT activation [10] |
| SH2 Domain-Binding Peptides | GpYLPQTV | STAT3 SH2 domain binding motif for competition assays | Fluorescence polarization assays [6] |
| Monobody Inhibitors | Mb(SHP2PTP_13) | Selective binding to SHP2 phosphatase domain | Molecular dynamics studies of inhibition [81] |
| Peptide Inhibitors | CSIP with l-OMT | C-SH2 domain targeting with non-hydrolysable pTyr mimetic | Selective disruption of SHP2 interactions [80] |
| Computational Tools | Molecular Dynamics Simulations | Analysis of binding stability and conformational dynamics | Mechanism of selective SHP2 inhibition [81] |
| Genetic Engineering | CRISPR/Cas9 Base Editing | Introduction of specific point mutations in SH2 domains | Disease modeling of STAT5B mutations [79] |
The targeting of SH2 domains represents a promising frontier in therapeutic development, offering opportunities for achieving exceptional selectivity in modulating signaling pathways. The STAT-SH2 domain in particular serves as a critical node controlling dimerization and transcriptional activity, with growing evidence supporting its druggability through multiple approaches. While challenges remain – including achieving cellular permeability for small molecules and ensuring functional specificity – recent advances in inhibitor design, biosensor technology, and computational modeling are accelerating progress in this field.
The emergence of first-in-class BTK SH2 domain inhibitors with unprecedented selectivity profiles demonstrates the considerable potential of this targeting strategy [78]. Similarly, the development of novel STAT3 SH2 inhibitors that effectively disrupt both phosphorylated and unphosphorylated STAT3 dimerization opens new avenues for therapeutic intervention in STAT3-driven cancers [6]. As our understanding of non-canonical SH2 domain functions in lipid binding and phase separation continues to expand [8], so too will opportunities for developing innovative therapeutic strategies that target these diverse activities. The continued refinement of research tools – from genetically encoded biosensors to advanced computational methods – will further illuminate SH2 domain biology and accelerate the development of next-generation therapeutics targeting these critical signaling modules.
Comparative Analysis of STAT3 vs. STAT5B SH2 Domain Mutations and Pathologies
The Signal Transducer and Activator of Transcription (STAT) family of proteins represents a critical node in cellular signaling, converting extracellular cytokine and growth factor signals into transcriptional responses within the nucleus. The Src Homology 2 (SH2) domain is the architectural centerpiece of STAT function, mediating both receptor recruitment and the fundamental step of STAT dimerization via reciprocal phosphotyrosine-SH2 interactions [5] [82]. This domain, consisting of a central anti-parallel β-sheet flanked by two α-helices in an αβββα motif, contains two functionally critical subpockets: the phosphotyrosine (pY) binding pocket and the pY+3 specificity pocket [5]. STAT activation culminates in the phosphorylation of a critical C-terminal tyrosine residue, enabling SH2 domain-mediated formation of transcriptionally active homo- or heterodimers that translocate to the nucleus [5] [83]. The integrity of the SH2 domain is therefore paramount to normal STAT function, and its disruption represents a key mechanism of disease pathogenesis. This review provides a comparative analysis of mutations within the SH2 domains of STAT3 and STAT5B, framing their distinct pathological impacts within the broader context of STAT dimerization research.
Structural and Functional Divergence in STAT3 and STAT5B SH2 Domains
Despite sharing a conserved core structure, STAT3 and STAT5B SH2 domains exhibit distinct features that influence their mutational landscape and functional outcomes. STAT-type SH2 domains are distinguished from Src-type domains by the presence of a C-terminal α-helix (αB') instead of a β-sheet [5]. This region, part of the evolutionary active region (EAR), contributes to the unique functional properties of STAT SH2 domains. Molecular dynamics studies reveal that STAT SH2 domains display significant flexibility, particularly in the accessible volume of the pY pocket, a critical consideration for drug discovery [5].
A key difference lies in their dimerization stability and DNA binding affinity. STAT5B exhibits a shorter half-life of phosphorylated dimers compared to STAT3, potentially due to weaker SH2-phosphotyrosine interactions [44]. This inherent difference may explain why certain STAT5B mutations (e.g., N642H) produce a more dramatic gain-of-function (GOF) by substantially stabilizing the active dimeric state, whereas analogous STAT3 mutations yield more moderate effects. The structural context of the SH2 domain thus creates distinct evolutionary constraints, resulting in different mutational hotspots and functional consequences for STAT3 versus STAT5B.
Pathological Spectrum of SH2 Domain Mutations
STAT3 Mutations: Immunodeficiency and Cancer Duality
STAT3 SH2 domain mutations demonstrate a remarkable duality, causing both loss-of-function (LOF) in immunodeficiency and GOF in oncogenic contexts. Germline heterozygous LOF mutations are the established cause of Autosomal-Dominant Hyper-IgE Syndrome (AD-HIES), characterized by recurrent infections, eczema, and skeletal abnormalities [5] [84]. The pathognomonic laboratory finding is elevated IgE, stemming from impaired Th17 differentiation and diminished IL-17/IL-22 production [5]. Notably, mouse models harboring human AD-HIES-associated STAT3 mutations sometimes fail to recapitulate the hyper-IgE phenotype, highlighting important species-specific differences in STAT3 immune regulation [84].
Conversely, somatic GOF mutations in the STAT3 SH2 domain drive several hematologic malignancies. The most frequent hotspot, particularly at position Y640, is strongly associated with T-cell Large Granular Lymphocytic Leukemia (T-LGLL) and NK/T-cell lymphomas [85] [86]. These mutations promote constitutive STAT3 dimerization and nuclear translocation, leading to persistent transcription of target genes that enhance cell survival and proliferation, such as BCL-XL and MCL-1 [5] [83] [85].
Table 1: Spectrum and Clinical Impact of STAT3 SH2 Domain Mutations
| Mutation | Location | Pathology | Type | Functional Impact | Key References |
|---|---|---|---|---|---|
| K591E/M, R609G, S611N, S614R | αA2, βB5, βB7, BC3 | AD-HIES, T-LGLL, Lymphomas | Germline (AD-HIES) & Somatic (Cancer) | LOF (AD-HIES) / GOF (Cancer) | [5] |
| Y640F | SH2 Domain | T-LGLL, CLPD-NK, Lymphomas | Somatic | GOF | [85] [86] |
| D661Y | SH2 Domain | γδ-T-cell Lymphomas | Somatic | GOF | [85] |
STAT5B Mutations: From Growth Defects to Leukemogenesis
STAT5B mutations similarly span a wide clinical spectrum but with distinct pathological emphasis. Germline LOF mutations cause growth hormone insensitivity and immune dysregulation, manifesting as short stature and immunodeficiency [44]. In stark contrast, somatic GOF mutations are potent drivers of hematologic malignancies, with a striking predilection for T-cell neoplasms.
The N642H mutation represents the most frequent STAT5B hotspot, particularly prevalent in γδ-T-cell lymphomas and enteropathy-associated T-cell lymphoma (EATL) type II [85]. Another critical residue, Y665, can be substituted to phenylalanine (Y665F) or histidine (Y665H), resulting in dramatically divergent functional consequences despite their proximity. Y665F is a clear GOF mutation associated with T-LGLL and T-cell prolymphocytic leukemia (T-PLL), while Y665H behaves as a LOF variant [44]. This exquisite sensitivity to specific amino acid changes underscores the delicate structural balance within the SH2 domain.
Table 2: Spectrum and Clinical Impact of STAT5B SH2 Domain Mutations
| Mutation | Location | Pathology | Type | Functional Impact | Key References |
|---|---|---|---|---|---|
| N642H | SH2 Domain | γδ-T-cell Lymphomas, EATL Type II, T-LGLL | Somatic | GOF | [44] [85] |
| Y665F | SH2 Domain | T-LGLL, T-PLL | Somatic | GOF | [44] [85] |
| Y665H | SH2 Domain | T-PLL (Single Case) | Somatic | LOF | [44] |
| Various LOF Mutations | SH2 Domain | Growth Hormone Insensitivity, Immune Dysregulation | Germline | LOF | [44] |
Comparative Mutation Frequencies and Clinical Correlations
The distribution and clinical impact of SH2 domain mutations differ significantly between STAT3 and STAT5B. STAT3 mutations are more frequently identified across a broader range of lymphoid malignancies, including T-LGLL, CLPD-NK, and various NK/T-cell lymphomas, with reported frequencies of 21-73% in T-LGLL and 13-70% in CLPD-NK [86]. STAT5B mutations, while less common overall, demonstrate a remarkable specificity for γδ-T-cell lineage malignancies, with N642H present in approximately 33-36% of γδ-PTCL and EATL type II cases [85].
Clinically, STAT3-mutated T-LGLL/CLPD-NK is associated with more aggressive disease features, including a higher incidence of neutropenia, severe neutropenia, and an increased requirement for therapeutic intervention, leading to a shorter time-to-therapy compared to wild-type cases [86].
Molecular Mechanisms of Pathogenesis
Dysregulation of Dimerization and Stability
SH2 domain mutations primarily exert their effects by altering the thermodynamics of STAT dimerization. GOF mutations, such as STAT5B-N642H, enhance the binding affinity between the phosphorylated tyrosine of one STAT monomer and the SH2 domain of its partner. Molecular modeling and surface plasmon resonance studies confirm that the N642H substitution markedly increases this binding affinity, resulting in prolonged persistence of the phosphorylated STAT5B dimer and enhanced occupancy at target gene enhancers [85]. This stabilization translates to sustained transcriptional activity even in the absence of continuous cytokine stimulation.
Conversely, LOF mutations (e.g., many STAT3 AD-HIES mutations) disrupt critical interactions required for phosphotyrosine binding or SH2 domain structural integrity, impairing dimerization and nuclear accumulation [5]. The precise effect is mutation-specific, with some variants completely abrogating function while others merely reduce transcriptional activity.
Alteration of Immune and Oncogenic Gene Programs
The functional impact of SH2 domain mutations manifests through the rewiring of specific gene expression programs. In primary T-cells, the STAT5B-Y665F GOF mutation drives the accumulation of CD8+ effector/memory and CD4+ regulatory T-cells, altering CD4+/CD8+ ratios, whereas the Y665H LOF mutation diminishes these populations [44]. At the molecular level, STAT5B GOF mutants upregulate known oncogenic targets including IL2Rα, BCL-XL, BCL2, MIR155HG, and HIF2α, promoting cell survival and proliferation [85]. Chromatin immunoprecipitation studies demonstrate markedly increased binding of mutants like N642H to the regulatory regions of these genes [85].
Experimental Methodologies for Functional Characterization
In Silico Modeling and Biophysical Analysis
Computational approaches provide the first insights into mutation impact. Molecular dynamics simulations model the effect of mutations on SH2 domain flexibility and pocket accessibility [5]. Free energy calculations predict changes in dimerization affinity, successfully differentiating between GOF and LOF variants, as demonstrated for STAT5B Y665F versus Y665H [44]. Surface plasmon resonance (SPR) directly quantifies binding affinity changes by measuring the interaction kinetics between phosphopeptides and purified mutant SH2 domains, confirming the enhanced affinity of STAT5B-N642H for its phosphotyrosine counterpart [85].
Experimental Workflow for STAT SH2 Domain Mutation Analysis
Cell-Based and Biochemical Assays
Lentiviral transduction introduces mutant STAT genes into cell lines (e.g., KAI3 NK cells) or primary human NK/T-cells to assess functional impact [85]. Phospho-STAT levels are quantified via western blotting using phospho-specific antibodies (e.g., pY705-STAT3, pY699-STAT5B), with GOF mutants typically showing sustained phosphorylation [85]. Transcriptional activity is evaluated through qPCR or RNA-Seq measurement of known STAT targets (BCL2, BCL-XL, MCL-1) and chromatin immunoprecipitation (ChIP) assays assessing DNA binding occupancy [44] [85]. Cellular proliferation and survival assays under limiting cytokine conditions demonstrate the growth advantage conferred by GOF mutations [85].
In Vivo Modeling
Transgenic "knock-in" mouse models, such as those expressing Stat5b-Y665F, provide physiological context by revealing impacts on immune cell populations, CD4+/CD8+ ratios, and disease phenotypes in a whole-organism setting [44]. These models are crucial for validating in vitro findings and understanding the complex pathophysiology of STAT mutations.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Investigating STAT SH2 Domain Mutations
| Reagent/Category | Specific Examples | Function & Application | Research Context |
|---|---|---|---|
| Phospho-Specific Antibodies | anti-pY705-STAT3, anti-pY699-STAT5B | Detects activated, phosphorylated STATs; Western blot, Flow Cytometry | [85] |
| JAK/STAT Inhibitors | JAK1/2 Inhibitors (e.g., Ruxolitinib) | Functional validation; tests pathway dependency and therapeutic potential | [85] |
| Lentiviral Expression Systems | STAT3/5B WT and mutant constructs, shRNA | Gene delivery for overexpression or knockdown in cell lines/primary cells | [44] [85] |
| Model Systems | KAI3 (NK-cell line), Primary Human NK/T-cells, Transgenic Mice | Cellular and in vivo models for functional studies | [44] [85] |
| Structural Biology Kits | Crystallization screens, SPR chips | Protein-protein interaction analysis and 3D structure determination | [5] [85] |
Therapeutic Implications and Future Directions
The central role of SH2 domains in STAT dimerization makes them attractive therapeutic targets. Current strategies include JAK inhibitors (e.g., Ruxolitinib) that indirectly suppress STAT activation upstream, which have shown partial efficacy against mutant STAT-driven growth in experimental models [82] [85]. Direct STAT inhibitors are under active development, including small molecules designed to block SH2 domain-mediated dimerization or DNA binding [5] [82]. Notably, STAT3 degraders based on proteolysis-targeting chimera (PROTAC) technology represent a promising new modality to directly eliminate the STAT3 protein [87].
Future research priorities include resolving high-resolution structures of full receptor-JAK-STAT complexes to inform rational drug design, developing mutant-specific inhibitors that spare wild-type STAT function to reduce toxicity, and translating insights from structural biology into clinical candidates that can effectively target the STAT SH2 domain in human diseases [5] [82].
The SH2 domain represents the functional core of STAT proteins, governing the critical dimerization step that defines their transcriptional activity. STAT3 and STAT5B SH2 domain mutations drive diverse pathologies—from profound immunodeficiency to aggressive hematologic malignancies—through opposing effects on protein function. While STAT3 mutations demonstrate a wider association with lymphoid malignancies, STAT5B mutations show remarkable lineage specificity for γδ-T-cell diseases. The molecular pathophysiology of these mutations centers on altered dimerization kinetics and stability, rewiring transcriptional programs that control immune function and cell survival. Ongoing research into the structural biology of these domains continues to inform therapeutic strategies aimed at directly targeting STAT dimerization, holding promise for more effective treatments for STAT-driven diseases.
The signal transducers and activators of transcription (STAT) family of proteins function as critical mediators of cellular signaling, with their dimerization status representing a fundamental regulatory node in transcriptional activation. While phosphotyrosine-SH2 domain interactions in activated STATs have been extensively characterized, the assembly and functional significance of latent unphosphorylated STAT (U-STAT) dimers remain less understood. This technical review provides a comprehensive assessment of U-STAT dimerization across the STAT family, examining the structural bases and functional implications of these complexes. We synthesize recent advances in characterizing U-STAT dimers through co-localization assays, structural analyses, and biophysical approaches, highlighting the central role of the SH2 domain in both phosphorylated and unphosphorylated STAT complexes. The findings presented herein reshape our understanding of STAT regulation and offer new perspectives for therapeutic intervention in STAT-driven pathologies.
The Janus kinase-signal transducer and activator of transcription (JAK-STAT) pathway serves as a central communication hub in cellular function, transmitting signals from more than 50 cytokines and growth factors [33]. STAT proteins are multidomain transcription factors that convert extracellular signals into transcriptional responses through a sophisticated mechanism involving phosphorylation, dimerization, and nuclear translocation [5] [33]. While the canonical activation pathway involves tyrosine phosphorylation and SH2 domain-mediated parallel dimerization, emerging evidence demonstrates that unphosphorylated STATs form distinct dimeric complexes with significant biological functions [88] [89].
The Src Homology 2 (SH2) domain is an evolutionarily conserved protein module that arose within metazoan signaling pathways approximately 600 million years ago [5]. In STAT proteins, this domain typically facilitates phosphotyrosine-mediated dimerization following activation. However, recent structural and functional studies have revealed that STAT SH2 domains participate in more complex regulatory networks than previously appreciated, including the formation of latent unphosphorylated dimers through alternative interfaces [88] [89] [90]. These U-STAT dimers exhibit distinct structural architectures compared to their phosphorylated counterparts and contribute to both canonical and non-canonical STAT functions.
This assessment systematically examines the landscape of latent STAT dimerization across the entire STAT family, with particular emphasis on the multifaceted roles of the SH2 domain. We integrate findings from structural biology, cellular assays, and biophysical approaches to provide a unified framework for understanding U-STAT complex formation and its implications for STAT regulation and function.
Structural Foundations of STAT Proteins and SH2 Domains
STAT Architecture and Domain Organization
STAT proteins share a conserved domain architecture consisting of six structural motifs: an amino-terminal domain (NTD), a coiled-coil domain (CCD), a DNA-binding domain (DBD), a linker domain (LD), an SH2 domain, and a carboxy-terminal transactivation domain (TAD) [42]. The SH2 domain serves as the critical mediator of protein-protein interactions, primarily through recognition of phosphotyrosine motifs. In canonical STAT activation, SH2 domains facilitate both receptor recruitment and STAT dimerization via reciprocal phosphotyrosine-SH2 interactions [5] [32].
Unique Features of STAT-Type SH2 Domains
STAT-type SH2 domains possess distinctive structural characteristics that differentiate them from Src-type SH2 domains. The core SH2 domain structure consists of a central anti-parallel β-sheet (βB-βD strands) flanked by two α-helices (αA and αB) in an αβββα arrangement [5]. STAT-type SH2 domains contain a C-terminal α-helix (αB'), whereas Src-type domains feature additional β-strands (βE and βF) in this region. The STAT SH2 domain contains two primary binding pockets:
- pY pocket: The phosphate-binding pocket formed by the αA helix, BC loop, and one face of the central β-sheet that accommodates the phosphotyrosine residue
- pY+3 pocket: The specificity pocket created by the opposite face of the β-sheet, αB helix, and CD and BC* loops that determines peptide binding specificity [5]
These structural features enable STAT SH2 domains to participate in diverse interactions, including the formation of both phosphorylated and unphosphorylated dimers through distinct interfaces.
Table 1: Structural Features of STAT-Type SH2 Domains
| Structural Element | Description | Functional Role |
|---|---|---|
| Central β-sheet | Anti-parallel βB-βD strands | Partitions SH2 domain into pY and pY+3 pockets |
| αA helix | Flanks one side of β-sheet | Forms part of pY pocket for phosphotyrosine binding |
| αB helix | Flanks opposite side of β-sheet | Contributes to pY+3 pocket formation |
| αB' helix | C-terminal extension in STAT-type SH2 domains | Distinguishes STAT-type from Src-type SH2 domains |
| BC loop | Connects βB-βC strands | Forms part of pY pocket |
| pY pocket | Formed by αA helix, BC loop, and β-sheet | Binds phosphotyrosine residues |
| pY+3 pocket | Formed by β-sheet, αB helix, and CD/BC* loops | Determines binding specificity |
| Hydrophobic system | Cluster of non-polar residues at base of pY+3 pocket | Stabilizes β-sheet conformation and SH2 domain integrity |
Family-Wide Assessment of Latent STAT Dimers
Systematic Analysis of U-STAT Dimerization
A comprehensive family-wide assessment of latent STAT dimerization employed a co-localization-based assay in living cells to examine all 28 possible combinations of the seven unphosphorylated STAT proteins [88]. This systematic approach revealed five U-STAT homodimers (STAT1, STAT3, STAT4, STAT5A, and STAT5B) and two heterodimers (STAT1:STAT2 and STAT5A:STAT5B), while STAT6 was found to be predominantly monomeric in its unphosphorylated state [88].
The experimental design utilized nucleocytoplasmic shuttling properties of STATs by engineering bait proteins with transferable nuclear localization signals (NLS) or nuclear export signals (NES). When co-expressed with untagged STATs, dimerization resulted in co-localization to the bait's compartment, enabling detection of interactions. This approach was validated using established STAT1 and STAT3 homodimers and appropriate dimer-disrupting mutants (STAT1-F77A and STAT3-L78R) [88].
Table 2: Latent STAT Dimerization Profile Across STAT Family
| STAT Protein | Homodimerization | Heterodimerization Partners | Structural Basis |
|---|---|---|---|
| STAT1 | Yes | STAT2 | N-domain mediated antiparallel dimer |
| STAT2 | No | STAT1 | Requires STAT1 for dimerization |
| STAT3 | Yes | Not detected in U-STAT | N-domain mediated antiparallel dimer |
| STAT4 | Yes | Not detected in U-STAT | N-domain mediated antiparallel dimer |
| STAT5A | Yes | STAT5B | N-domain mediated antiparallel dimer |
| STAT5B | Yes | STAT5A | N-domain mediated antiparallel dimer |
| STAT6 | No | None detected | Monomeric in unphosphorylated state |
Structural Bases of Unphosphorylated STAT Dimers
Structural studies have revealed that unphosphorylated STAT dimers adopt architectures distinct from their phosphorylated counterparts. Crystallographic analysis of unphosphorylated STAT1 demonstrated two dimer interfaces: one between N-terminal domains (ND) and another between core fragments (CF) [90]. The connector region between ND and CF provides flexibility, allowing interconversion between antiparallel and parallel orientations of the core fragments as determined by SH2 domain positioning [90].
Small-angle X-ray scattering (SAXS) studies of unphosphorylated STAT5a have confirmed the coexistence of monomeric and dimeric species in solution, with the dimer corresponding to an antiparallel arrangement [89]. The dissociation constant for STAT5a core domain dimerization was determined to be Kd ∼ 90 μM, significantly weaker than full-length protein dimerization, suggesting a complex network of intermolecular interactions of varying affinities regulates STAT oligomerization [89].
Diagram 1: STAT Dimerization States and Transitions. Unphosphorylated STATs exist in equilibrium between monomeric and antiparallel dimeric states, with dimerization mediated primarily by N-domain interactions. Upon phosphorylation, STATs undergo structural rearrangement to form parallel dimers stabilized by reciprocal SH2-phosphotyrosine interactions.
Experimental Approaches for Assessing Latent STAT Dimers
Co-Localization Assay in Living Cells
The co-localization assay provides a robust method for detecting U-STAT dimerization in living cells [88]. This approach leverages the nucleocytoplasmic shuttling capability of STAT proteins and can be implemented with the following protocol:
Experimental Workflow:
- Engineer bait STAT constructs fused with strong heterologous NLS or NES signals
- Co-express bait constructs with untagged test STAT proteins in appropriate cell lines
- Monitor subcellular localization using fluorescent protein tags (e.g., mEGFP, mCherry)
- Quantify co-localization by calculating fluorescence distribution ratios between compartments
- Validate interactions using known dimer-disrupting mutants (e.g., STAT1-F77A, STAT3-L78R) as negative controls
Critical Considerations:
- Maintain bait expression within 1-4-fold excess over test protein concentration
- Include appropriate controls for proper NLS/NES function
- Account for cell-to-cell variability by analyzing multiple cells across independent experiments
- Verify expression of full-length fusion proteins by Western blotting
Biophysical and Structural Approaches
Multiple biophysical techniques have been employed to characterize U-STAT dimers:
Small-Angle X-Ray Scattering (SAXS):
- Enables structural characterization of proteins in solution under near-native conditions
- Provides parameters including radius of gyration (Rg) and maximum particle distance (Dmax)
- Allows monitoring of concentration-dependent oligomeric equilibria
- Successfully applied to identify monomer-dimer equilibrium of STAT5a (Kd ∼ 90 μM) [89]
Analytical Ultracentrifugation:
- Determines molecular weights and association constants in solution
- Previously used to establish high-affinity homodimerization of unphosphorylated STAT1 (Kd in low nM range) [88]
Crystallography:
- Provides atomic-resolution structures of STAT complexes
- Revealed antiparallel arrangement of unphosphorylated STAT1 dimers [90]
- Identified parallel arrangement with interacting SH2 domains in unphosphorylated STAT3 [89]
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Studying Latent STAT Dimers
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Expression Constructs | STAT-NLS, STAT-NES baits | Directed localization for co-localization assays |
| Fluorescent Tags | mEGFP, mCherry | Visualizing subcellular localization and protein interactions |
| Dimer-Disrupting Mutants | STAT1-F77A, STAT3-L78R | Negative controls for dimerization assays |
| Cell Lines | HEK 293T, EPT3M1-STAT3 | Model systems for STAT expression and functional studies |
| Detection Assays | Co-immunoprecipitation, Western blot | Validating protein interactions and expression |
| SH2 Domain Inhibitors | S3I-201, 323-1, 323-2 (delavatine A derivatives) | Probing SH2 domain function in dimerization [42] |
| Cytokines/Growth Factors | IL-6, IFN-γ, EGF | STAT activation controls and pathway stimulation |
Functional Implications of Latent STAT Dimers
Roles in Cellular Signaling and Transcription
Latent STAT dimers fulfill critical functions beyond serving as precursors for activated STATs:
Prerequisite for Activation:
- U-STAT4 homodimerization is required for cytokine-induced STAT4 activation [88]
- U-STAT1:STAT2 heterodimerization modulates type I and type II interferon signaling [88]
Transcriptional Regulation:
- Unphosphorylated STATs participate in transcription and gene regulation independently of tyrosine phosphorylation [89]
- U-STAT3 can drive and coregulate transcription and function as a microtubule stabilizer in the cytoplasm [89]
Cellular Homeostasis:
- Unphosphorylated STATs continuously shuttle between cytoplasm and nucleus [89]
- Contribute to basal gene regulation and rapid response capabilities
Pathological Relevance and Therapeutic Targeting
Dysregulation of STAT dimerization is implicated in various diseases, with the SH2 domain representing a hotspot for mutations in pathological conditions [5]. STAT3 SH2 domain mutations have been identified in multiple pathologies:
- Autoimmune Disease: AD-HIES (autosomal-dominant hyper IgE syndrome) mutations (K591E, K591M, R593P, S611N) cause loss-of-function [5]
- Leukemias/Lymphomas: S614R mutation in T-LGLL, NK-LGLL, ALK-ALCL, and HSTL leads to gain-of-function [5]
- Solid Tumors: E616K mutation in NKTL and E616G in DLBCL demonstrate oncogenic potential [5]
The therapeutic potential of targeting STAT SH2 domains is evidenced by developing small molecule inhibitors such as 323-1 and 323-2 (delavatine A stereoisomers), which directly target the STAT3 SH2 domain and inhibit both phosphorylated and non-phosphorylated STAT3 dimerization [42]. These compounds bind to three subpockets of the STAT3 SH2 domain and demonstrate greater potency than the reference inhibitor S3I-201 [42].
Diagram 2: STAT SH2 Domain in Health and Disease. The STAT SH2 domain serves as both a mutation hotspot in disease and a therapeutic target. Different mutations can lead to either loss-of-function (as in AD-HIES) or gain-of-function (as in various cancers), while small molecule inhibitors targeting this domain can modulate dimerization and activity for therapeutic benefit.
This comprehensive assessment of latent STAT dimers reveals a sophisticated regulatory landscape in which unphosphorylated STAT complexes play integral roles in cellular signaling and homeostasis. The systematic characterization of U-STAT dimers across the STAT family demonstrates considerable structural and functional diversity in the mechanisms linking STAT dimerization before and after activation. The SH2 domain emerges as a central player in both phosphorylated and unphosphorylated STAT complexes, with its multifaceted functions extending beyond canonical phosphotyrosine recognition to include participation in alternative dimer interfaces and regulatory interactions.
These insights fundamentally advance our understanding of STAT biology and open new avenues for therapeutic intervention. The development of small molecule inhibitors targeting STAT SH2 domains, particularly those capable of disrupting both phosphorylated and unphosphorylated dimers, represents a promising strategy for modulating pathological STAT signaling in cancer and inflammatory diseases. Future research elucidating the structural dynamics and functional consequences of latent STAT dimers will continue to refine our understanding of this critical signaling pathway and its therapeutic potential.
The Janus kinase-signal transducer and activator of transcription (JAK-STAT) pathway represents a fundamental signaling mechanism employed by cytokines, growth factors, and hormones to reprogram cellular gene expression. For decades, the paradigm of STAT signaling centered on cytokine-induced phosphorylation of monomeric STATs, leading to the formation of canonical homodimers via reciprocal phosphotyrosine-SH2 domain interactions. These homodimers then translocate to the nucleus to drive the transcription of specific target genes. However, a significant conundrum has persisted in cytokine biology: while over 50 cytokines play distinct immunological roles, they must transmit these complex signals using only seven STAT molecules [91]. This discrepancy suggests additional layers of signaling complexity.
We now understand that STAT heterodimerization represents a crucial mechanism diversifying cytokine signals. These heterodimers—comprising two different STAT proteins—exhibit unique DNA-binding specificities, target gene preferences, and functional outcomes beyond those of their constituent homodimers. The SH2 domain, a structurally conserved protein interaction module, is central to both canonical homodimer and non-canonical heterodimer formation. This technical guide examines the validated landscape of STAT heterodimers, details methodologies for their study, and explores their implications for therapeutic development, all within the critical context of SH2 domain-mediated interactions.
STAT Heterodimer Landscape and Formation Mechanisms
Known STAT Heterodimers and Their Activators
STAT heterodimers are not rare exceptions but rather integral components of the cytokine signaling network. Systematic assessments using co-localization assays in living cells have identified specific heterodimers among unphosphorylated STATs (U-STATs), including STAT1:STAT2 and STAT5A:STAT5B [88]. Importantly, cytokine stimulation induces additional heterodimer combinations. The table below summarizes key STAT heterodimers, their activating cytokines, and potential functional roles.
Table 1: Characterized STAT Heterodimers and Their Activators
| Heterodimer Pair | Activating Cytokines | Functional Context |
|---|---|---|
| STAT1:STAT2 | Type I Interferons [91] | Forms ISGF3 complex with IRF9; binds ISRE elements [91] |
| STAT1:STAT3 | IL-6, IL-10, IL-27 [91] | May integrate inflammatory/anti-inflammatory signals |
| STAT1:STAT4 | IL-35 [91] | Associated with regulatory T-cell function |
| STAT3:STAT5 | IL-2, IL-7, G-CSF, M-CSF [91] | Links survival/growth factor signaling |
| STAT5A:STAT5B | Constitutively formed as U-STATs [88] | Forms latent cytoplasmic complexes |
Structural Basis of Heterodimer Formation
The formation of STAT heterodimers is governed by precise structural mechanisms, with the SH2 domain playing the pivotal role.
- Phosphotyrosine-SH2 Interactions: Canonical active dimers (both homo- and heterodimers) are stabilized by reciprocal phosphotyrosine-SH2 domain interactions, where the phosphorylated tyrosine (pY701 in STAT1) of one monomer engages the SH2 domain of its partner [92].
- Requirement for Tyrosine Phosphorylation: The formation of cytokine-induced STAT1 heterodimers strictly depends on prior tyrosine phosphorylation. Mutational studies demonstrate that STAT1 with a defective SH2 domain (R602K) or mutated phosphorylation site (Y701F) fails to undergo serine phosphorylation or proper heterodimerization in response to IFN-γ, though it still responds to stress signals like UV irradiation [92].
- Latent Unphosphorylated Dimers: Before activation, certain STATs pre-form latent heterodimers in an unphosphorylated state. These U-STAT heterodimers, such as STAT1:STAT2, are stabilized by N-terminal domain interactions rather than phosphotyrosine-SH2 contacts, creating an antiparallel conformation distinct from the parallel orientation of active dimers [88]. This pre-dimerization can facilitate rapid activation upon cytokine stimulation.
Experimental Validation of STAT Heterodimers
Methodologies for Detection and Validation
Nucleocytoplasmic Co-localization Assay
A powerful approach for detecting latent U-STAT heterodimers in live cells involves a co-localization assay that exploits the nucleocytoplasmic shuttling properties of STATs [88].
Experimental Workflow: Nucleocytoplasmic Co-localization Assay
Protocol Details:
- Bait Protein Engineering: Fuse the STAT of interest to well-characterized nuclear localization signals (NLS, e.g., from SV40 T-antigen) or nuclear export signals (NES, e.g., from protein kinase A inhibitor) [88].
- Test Protein Preparation: Fuse candidate interacting STATs with fluorescent tags (e.g., mEGFP, mCherry). These typically display pancellular distribution when expressed alone.
- Cell Transfection and Imaging: Co-express bait and test proteins in appropriate cell lines (e.g., STAT-deficient fibroblasts). Quantify co-localization using fluorescence microscopy.
- Controls and Validation: Include monomeric STAT mutants (e.g., STAT1-F77A, STAT3-L78R) as negative controls. These mutants contain point mutations in their N-terminal dimerization interfaces that disrupt latent dimer formation without affecting nucleocytoplasmic shuttling [88].
FRET-Based Biosensors for Active Dimers
Genetically encoded biosensors using Förster resonance energy transfer (FRET) detected by fluorescence lifetime imaging microscopy (FLIM) enable real-time monitoring of STAT activation and heterodimerization in live cells [10].
STATeLight Biosensor Engineering:
- Fluorophore Selection: Tag STAT monomers with appropriate FRET pairs (e.g., mNeonGreen donor and mScarlet-I acceptor) [10].
- Optimal Tagging Positions: C-terminal fusion to the SH2 domain provides the highest FRET efficiency change upon activation, as this positioning places fluorophores in close proximity during parallel dimer formation [10].
- Activation Readout: Upon cytokine stimulation, conformational change from antiparallel to parallel dimers decreases donor fluorescence lifetime, indicating heterodimer formation.
Figure 2: FRET-FLIM Biosensor for STAT Dimerization
Specificity Controls and Validation
Rigorous validation is essential to confirm heterodimer specificity:
- SH2 Domain Mutants: Test STAT variants with defective SH2 domains (e.g., R602K in STAT1) to confirm phosphorylation-dependent heterodimerization [92].
- Expression Level Controls: Ensure bait and test proteins are expressed at appropriate ratios (equimolar to 4:1 test:bait) to avoid non-specific crowding effects [88].
- Competition Experiments: Validate heterodimer specificity using purified SH2 domains in competitive binding assays with phosphopeptide libraries [93] [94].
Functional Specificity of STAT Heterodimers
Distinct Gene Regulatory Profiles
STAT heterodimers exhibit unique transcriptional programs distinct from their homodimeric counterparts. This functional specificity arises through several mechanisms:
- DNA Binding Specificity: Heterodimers recognize variations of the gamma-activated sequence (GAS) elements. For instance, STAT1:STAT2 heterodimers bind distinct consensus sequences in a subset of interferon-stimulated genes compared to STAT1 or STAT2 homodimers [91].
- Differential Serine Phosphorylation Requirements: The C-terminal transactivation domains of STATs contain conserved serine residues (e.g., S727 in STAT1) whose phosphorylation differentially affects target genes. The STAT1 S727A mutation strongly reduces induction of certain genes (e.g., GBP1, TAP1) while minimally affecting others, suggesting gene-specific requirements for serine phosphorylation that likely influence heterodimer function [92].
- Promoter-Specific Cooperation: Heterodimers recruit distinct coactivator complexes compared to homodimers, leading to altered transcriptional outcomes at specific promoters.
Biological Consequences in Immunity and Disease
STAT heterodimers expand the functional repertoire of cytokine signaling in crucial biological contexts:
- Immune Cell Differentiation: Heterodimers help specify T-helper cell differentiation. For example, STAT1:STAT4 heterodimers induced by IL-35 contribute to regulatory T-cell function, while STAT3:STAT4 heterodimers may integrate IL-23 and IL-12 signaling during Th17 differentiation [91].
- Signal Modulation: STAT1:STAT2 heterodimers have positive or negative consequences for type 1 and type 2 interferon signaling, respectively, demonstrating how heterodimerization can either enhance or dampen specific cytokine responses [88].
- Disease Implications: Aberrant STAT heterodimer formation contributes to pathogenesis. Unphosphorylated STAT3 (U-STAT3) forms dimers that regulate distinct genes involved in tumorigenesis, and heterodimer-specific signaling has been implicated in cancer, autoimmunity, and immunodeficiency [4] [10].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for STAT Heterodimer Research
| Reagent / Tool | Function & Utility | Key Applications |
|---|---|---|
| STATeLight FRET Biosensors [10] | Genetically encoded sensors (FP-tagged STATs) for real-time dimer detection | Live-cell imaging of STAT activation kinetics; compound screening |
| NLS/NES-STAT Bait Proteins [88] | Engineered STATs with strong localization signals | Detection of latent U-STAT heterodimers via co-localization assays |
| SH2 Domain Mutants (e.g., STAT1-R602K) [92] | Defective in phosphotyrosine binding | Controls for phosphorylation-dependent dimerization mechanisms |
| Monomeric STAT Mutants (e.g., STAT1-F77A, STAT3-L78R) [88] | Disrupted N-terminal dimerization interface | Negative controls for specific dimer interactions |
| Computational Prediction Tools (e.g., GalaxyHeteromer) [95] | Template-based and ab initio heterodimer structure prediction | In silico modeling of STAT heterodimer interfaces and stability |
| Selective Kinase Inhibitors | Probe serine phosphorylation requirements (e.g., p38MAPK inhibitors) [92] | Dissecting signaling pathways leading to heterodimer post-translational modifications |
The study of STAT heterodimers represents a paradigm shift in understanding JAK-STAT signaling complexity. Moving beyond the canonical homodimer model reveals a sophisticated regulatory network where limited STAT molecules achieve remarkable signaling specificity through combinatorial heterodimerization. The SH2 domain serves as the central architectural element enabling this diversity, with its specificity for phosphotyrosine motifs determining partner selection and functional outcomes.
Future research must focus on developing heterodimer-selective probes and inhibitors that can distinguish between different STAT combinations, much like the bivalent ligands successfully engineered for GPCR heterodimers [96]. The integration of advanced biosensors [10], predictive computational models [95], and structural insights into SH2 domain specificity [93] [94] will accelerate this progress. As we continue to decipher the heterodimer code, we unlock new opportunities for precisely targeted therapeutic interventions in cancer, autoimmune diseases, and immune disorders where specific STAT heterodimers drive pathogenesis.
The Src homology 2 (SH2) domain is a critical protein interaction module that specifically recognizes and binds to phosphotyrosine (pY) motifs, thereby orchestrating a vast network of cellular signaling pathways. In the context of the Signal Transducer and Activator of Transcription (STAT) family, the SH2 domain plays an indispensable role in mediating STAT dimerization, a fundamental step for nuclear translocation and transcriptional activation. The canonical activation model involves phosphorylation of a key tyrosine residue (e.g., Y705 in STAT3), which then engages in reciprocal SH2-pY interaction with another STAT monomer to form a functional dimer. Given that aberrant activation of STAT proteins, particularly STAT3, is a hallmark of numerous cancers and autoimmune diseases, the STAT-SH2 interface presents a compelling therapeutic target. However, the high conservation of the phosphotyrosine-binding pocket across SH2 domains has historically made the development of selective orthosteric inhibitors exceptionally challenging. This whitepaper examines emerging strategies that circumvent these limitations through two complementary approaches: allosteric inhibition and targeted disruption of protein-protein interactions (PPIs), providing a technical guide for researchers and drug development professionals.
Structural and Mechanistic Basis of STAT-SH2 Function
Domain Architecture and Dimerization Mechanism
STAT proteins are multi-domain transcription factors with a conserved architecture. STAT3, for example, comprises an N-terminal domain (NTD), a coiled-coil domain (CCD), a DNA-binding domain (DBD), a linker domain (LD), the SH2 domain, and a transactivation domain (TAD). The SH2 domain is central to the classic activation cycle. Upon cytokine or growth factor stimulation, Janus kinases (JAKs) or other tyrosine kinases phosphorylate the conserved tyrosine residue (Y705). This phosphorylation event triggers a conformational change that enables the SH2 domain of one STAT monomer to bind the pY-containing peptide of another, forming an active dimer primarily through reciprocal SH2-pY705 interactions [97] [98]. These dimers then translocate to the nucleus, bind specific DNA response elements, and drive the expression of target genes involved in cell proliferation, survival, and immune regulation.
Allosteric Regulation of STAT3
Emerging evidence reveals that STAT proteins are subject to sophisticated allosteric regulation. Studies using NMR spectroscopy on STAT3 have demonstrated that binding of a phosphopeptide to the SH2 domain induces structural and dynamic changes in distal domains, including the linker domain (LD) and the DNA-binding domain (DBD). This inter-domain communication suggests that STAT3 functions as a dynamically allosteric protein [99]. For instance, a disease-associated mutation (I568F) in the linker domain was found to induce conformational changes in the SH2 domain and reduce its binding affinity for pY-peptides, without being directly involved in the binding interface. This discovery underscores the existence of a dynamic allosteric network connecting different STAT3 domains, thereby revealing novel potential sites for therapeutic intervention beyond the conserved orthosteric pocket [99].
The following diagram illustrates the key domains and the allosteric network within STAT3, highlighting how perturbations in one domain can affect distal sites.
Emerging Allosteric Targeting Strategies
Direct Allosteric Inhibition of STAT3
Directly targeting STAT3 via allosteric mechanisms has gained significant traction to overcome the selectivity challenges of orthosteric SH2 inhibitors. These strategies focus on domains outside the conserved pY-binding site.
- Targeting the Coiled-Coil Domain (CCD): The CCD represents a promising allosteric site. The compound K116 is a pioneering allosteric inhibitor identified to bind the STAT3 CCD. In breast cancer models, K116 inhibited cell proliferation in a dose-dependent manner by reducing STAT3 Y705 phosphorylation. It induced apoptosis, inhibited migration, and blocked pY705-STAT3 nuclear translocation. Furthermore, in a 4T1 murine breast cancer model, K116 (30 mg kg⁻¹) significantly suppressed tumor growth, providing in vivo validation for targeting the CCD [100].
- Targeting the N-Terminal Domain (NTD): The NTD is involved in stabilizing weak DNA binding and unphosphorylated dimer formation. While small molecules targeting the NTD are less common, protein-based inhibitors like monobodies (synthetic binding proteins) have been successfully developed. Monobodies such as MS3-N3 bind the NTD with high affinity (KD ≈ 40 nM) and have demonstrated exquisite selectivity for STAT3 over other STAT family members [98].
Table 1: Representative Direct Allosteric Inhibitors of STAT3
| Inhibitor | Target Domain | Mechanism of Action | Affinity/Potency | Stage |
|---|---|---|---|---|
| K116 [100] | Coiled-Coil (CCD) | Reduces pY705 phosphorylation, blocks nuclear translocation | IC₅₀ (cell proliferation): Low µM range; In vivo dose: 30 mg/kg | Preclinical |
| MS3-6 [98] | Coiled-Coil (CCD) | Bends CCD, impairing DNA binding & nuclear translocation; disrupts IL-22R interaction | KD = 7.6 nM | Research Tool |
| MS3-N3 [98] | N-Terminal (NTD) | Binds NTD, function potentially related to disrupting weak DNA binding or dimerization | KD = 40 nM | Research Tool |
Allosteric Inhibition of Upstream Regulators: SHP2 Phosphatase
Targeting upstream non-receptor tyrosine phosphatases that regulate STAT signaling is another viable allosteric strategy. SHP2, a phosphatase encoded by PTPN11, positively regulates the RAS/MAPK and other pathways and is a key node in oncogenic signaling.
- Allosteric SHP2 Inhibitors: SHP2 adopts an autoinhibited conformation where its N-SH2 domain blocks the catalytic PTP site. Allosteric inhibitors, such as SHP099, bind a tunnel-shaped pocket at the interface of the N-SH2, C-SH2, and PTP domains, stabilizing the autoinhibited state [101] [102]. This mechanism offers superior selectivity compared to catalytic site inhibitors.
- Next-Generation Inhibitors and Combinations: A structure-guided expansion strategy led to the development of novel allosteric inhibitors B1 and B8, based on a pyrrolo[2,1-f][1,2,4]triazin-4(3H)-one scaffold. These compounds exhibited enhanced potency with IC₅₀ values of 39 nM and 15 nM, respectively, acceptable pharmacokinetics, and potential oral bioavailability. Notably, B8 demonstrated profound synergy with the MCL-1 inhibitor VU661013 in acute myeloid leukemia (AML) models, unveiling a promising combinatorial approach [101].
Table 2: Representative Allosteric SHP2 Inhibitors
| Inhibitor | Mechanism of Action | Key Biochemical Properties | Development Stage |
|---|---|---|---|
| SHP099 [102] | Stabilizes autoinhibited conformation by binding tunnel site | First-generation allosteric inhibitor | Preclinical/Prototype |
| TNO155, RMC-4630 [102] | Stabilizes autoinhibited conformation | Clinical candidates | Phase I/II Trials |
| B1, B8 [101] | Structure-guided expansion of scaffold; synergistic with MCL-1 inhibition | IC₅₀ = 39 nM (B1), 15 nM (B8); acceptable PK | Preclinical |
The signaling pathway and allosteric inhibition mechanisms for SHP2 are summarized in the following diagram.
Experimental and Technical Approaches
This section details key methodologies for studying allosteric inhibition and disrupting STAT-SH2 interactions.
Key Experimental Protocols
Protocol 1: Cellular Thermal Shift Assay (CETSA) for Target Engagement
- Objective: To confirm direct binding of a small-molecule inhibitor (e.g., K116) to its intended protein target (e.g., STAT3) in a cellular context.
- Methodology:
- Cell Treatment: Treat cells (e.g., MDA-MB-468 breast cancer cells) with the inhibitor or vehicle control (DMSO).
- Heating: After incubation, harvest and wash cells. Divide the cell suspension into smaller aliquots and heat each at a gradient of temperatures (e.g., 34°C, 39°C, 44°C, 49°C, 54°C, 59°C) for 3 minutes.
- Lysis: Freeze-thaw cycles to lyse cells. Centrifuge at high speed (20,000g) to separate soluble protein from precipitated aggregates.
- Analysis: Analyze the soluble fraction by SDS-PAGE and Western blotting for the target protein.
- Data Interpretation: If the compound binds and stabilizes the target, the protein will remain soluble at higher temperatures in the treated samples compared to the control, observed as a shift in its thermal denaturation profile [100].
Protocol 2: Deep Mutational Scanning (DMS) for Functional Analysis of SHP2
- Objective: To comprehensively characterize the functional impact of thousands of mutations across a multi-domain protein like SHP2 on its phosphatase activity and regulation.
- Methodology:
- Library Construction: Create saturation mutagenesis libraries for full-length SHP2 (SHP2FL) and its isolated PTP domain (SHP2PTP) using methods like mutagenesis by integrated tiles (MITE).
- Yeast Selection: Co-express the SHP2 variant libraries with an active tyrosine kinase (v-Src or c-Src) in yeast. Yeast growth becomes dependent on SHP2's catalytic activity to counteract kinase-induced toxicity.
- Selection & Sequencing: Induce kinase and phosphatase expression, allow for outgrowth, and sequence the SHP2-coding DNA before and after selection to calculate enrichment scores for each variant.
- Data Interpretation: Variants with gain-of-function mutations are enriched after selection, while loss-of-function mutations are depleted. This reveals functional hotspots and provides insights into the pathogenicity of clinical variants and inter-domain allostery [46].
Protocol 3: Utilizing Monobodies for Targeted Protein Degradation
- Objective: To achieve selective degradation of an intracellular target protein (e.g., STAT3) and study the functional consequences.
- Methodology:
- Fusion Construct: Fuse a high-affinity, selective monobody (e.g., MS3-6) to the E3 ubiquitin ligase substrate receptor VHL, creating a monobody-VHL fusion.
- Cell Transfection: Stably or transiently express the monobody-VHL fusion in the target cell line.
- Induction of Degradation: The monobody engages the target protein (STAT3), and the VHL moiety recruits the endogenous ubiquitin-proteasome machinery, leading to polyubiquitination and degradation of STAT3.
- Validation: Monitor STAT3 protein levels over time by immunoblotting. Use a proteasome inhibitor (e.g., MG132) as a control to confirm the mechanism is proteasome-dependent [98].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Investigating STAT-SH2 and Allosteric Inhibition
| Reagent / Tool | Function / Target | Key Application |
|---|---|---|
| K116 [100] | Small molecule allosteric inhibitor of STAT3 CCD | Inhibiting STAT3 phosphorylation, nuclear translocation, and tumor growth in preclinical models. |
| SHP099 & Derivatives (B1, B8) [101] [102] | Small molecule allosteric inhibitors of SHP2 | Stabilizing SHP2 autoinhibition; studying RAS/MAPK/JAK-STAT signaling; combination therapy studies. |
| STAT3 Monobodies (MS3-6, MS3-N3) [98] | Synthetic binding proteins targeting STAT3 CCD/NTD | High-affinity, selective intracellular inhibition; mechanistic studies; fused to VHL for targeted degradation. |
| VHL Fusion System [98] | E3 ubiquitin ligase substrate receptor | Targeted degradation of proteins when fused to a target-binding agent (e.g., monobody, small molecule). |
| Phospho-specific STAT3 (Y705) Antibody | Detects activated STAT3 | Assessing STAT3 activation status by Western blot, immunofluorescence, and flow cytometry. |
| SHP2 Deep Mutational Scanning Library [46] | Comprehensive SHP2 mutant library | High-throughput functional profiling of SHP2 variants to identify regulatory mechanisms and pathogenic mutations. |
The therapeutic targeting of the STAT-SH2 domain and its associated signaling modules is undergoing a paradigm shift, moving beyond the challenging orthosteric pY-binding site. Strategies that leverage allosteric inhibition, as exemplified by CCD-targeting K116 and tunnel-binding SHP2 inhibitors, and those that employ innovative PPI disruptors like monobodies, are yielding compounds with enhanced selectivity and novel mechanisms. Critical to this progress are advanced experimental techniques, including CETSA for cellular target engagement, DMS for functional proteomics, and monobody-based degradation tools, which collectively empower researchers to decomplexify signaling networks and validate new drug targets. The promising synergistic efficacy observed in combinations, such as SHP2 and MCL-1 inhibition in AML, further highlights the potential of these strategies in overcoming resistance and improving therapeutic outcomes. As our understanding of allosteric networks and domain-domain communications within proteins like STAT3 and SHP2 deepens, the rational design of next-generation therapeutics targeting these critical oncogenic signaling nodes will continue to accelerate.
Conclusion
The STAT-SH2 domain is a central processing unit in cellular signaling, whose structural integrity is paramount for precise STAT dimerization and transcriptional control. The delicate evolutionary balance within this domain is underscored by the fact that mutations at identical residues can lead to either activating or deactivating pathologies, highlighting its exquisite sensitivity. While the canonical role of phosphotyrosine-mediated dimerization is well-established, emerging research on unphosphorylated STAT dimers and non-canonical functions expands the therapeutic landscape. Future directions must leverage advanced structural and dynamic studies to uncover convergent mechanisms of disease-associated mutations. The development of small-molecule inhibitors targeting the STAT-SH2 domain, though challenging, holds immense promise for precision medicine in oncology and immunology, offering potential to disrupt pathogenic signaling at its core.
References
- 1. SH2db - Contact [http://sh2db.ttk.hu/about]
- 2. Review The SH2 domain: versatile signaling module and ... [https://www.sciencedirect.com/science/article/abs/pii/S1570963904002882]
- 3. SH2 Domains Recognize Contextual Peptide Sequence ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2984226/]
- 4. Advances in research on unphosphorylated STAT3: A review [https://pmc.ncbi.nlm.nih.gov/articles/PMC12303475/]
- 5. Structural Implications of STAT3 and STAT5 SH2 Domain ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6895964/]
- 6. Novel STAT3 Inhibitors Targeting STAT3 Dimerization by ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2022.836724/full]
- 7. SH2db, an information system for the SH2 domain - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10320075/]
- 8. Update on Structure and Function of SH2 Domains [https://www.mdpi.com/1422-0067/26/18/9060]
- 9. Loops Govern SH2 Domain Specificity by Controlling Access ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6590088/]
- 10. Real-time visualization of STAT activation in live cells using ... [https://www.nature.com/articles/s41589-025-02012-0]
- 11. Update on Structure and Function of SH2 Domains [https://pmc.ncbi.nlm.nih.gov/articles/PMC12470777/]
- 12. SH2 Domains: Folding, Binding and Therapeutical ... [https://www.mdpi.com/1422-0067/23/24/15944]
- 13. Defining the Specificity Space of the Human Src Homology ... [https://www.sciencedirect.com/science/article/pii/S1535947620312007]
- 14. Phosphotyrosine recognition domains: the typical, the atypical ... [https://biosignaling.biomedcentral.com/articles/10.1186/1478-811X-10-32]
- 15. SH2 domain [https://en.wikipedia.org/wiki/SH2_domain]
- 16. Molecular Mechanisms of SH2- and PTB-Domain-Containing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3839611/]
- 17. The SH of 2 1 and domains 2 mediate multiple interactions in... Stat Stat [https://pubmed.ncbi.nlm.nih.gov/8605877/]
- 18. JAK- STAT signaling pathway - Wikipedia [https://en.wikipedia.org/wiki/JAK-STAT_signaling_pathway]
- 19. Surface Loops in a Single SH2 Domain Are Capable of ... [https://www.sciencedirect.com/science/article/pii/S1535947620318776]
- 20. Affinity, specificity, and kinetics of the interaction of the SHC ... [https://pubmed.ncbi.nlm.nih.gov/8550571/]
- 21. SH2 Domains Serve as Lipid-Binding Modules for pTyr ... [https://pubmed.ncbi.nlm.nih.gov/27052731/]
- 22. High-throughput profiling of sequence recognition by ... [https://elifesciences.org/articles/82345]
- 23. Structural Implications of STAT3 and STAT5 SH2 Domain ... [https://www.mdpi.com/2072-6694/11/11/1757]
- 24. Identification of the Linker-SH2 Domain of STAT as ... [https://www.sciencedirect.com/science/article/pii/S1535947620338639]
- 25. CoDIAC: A comprehensive approach for interaction ... [https://pubmed.ncbi.nlm.nih.gov/41252491/]
- 26. SH2 Domains: Folding, Binding and Therapeutical Approaches [https://pmc.ncbi.nlm.nih.gov/articles/PMC9783222/]
- 27. Analysis of the Fyn SH2 domain structure - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC2585567/]
- 28. Distal Loop Flexibility of a Regulatory Domain Modulates ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3764022/]
- 29. Btk SH2-kinase interface is critical for allosteric ... [https://www.nature.com/articles/s41467-020-16128-5]
- 30. Intramolecular hydrophobic interactions are critical ... [https://www.nature.com/articles/srep35454]
- 31. Correlation between binding and dynamics at SH2 domain ... [https://www.nature.com/articles/nsb0298-156]
- 32. Crystal Structure of a Tyrosine Phosphorylated STAT-1 ... [https://www.sciencedirect.com/science/article/pii/S0092867400814439]
- 33. The JAK/STAT signaling pathway: from bench to clinic [https://www.nature.com/articles/s41392-021-00791-1]
- 34. A practical guide to evaluating colocalization in biological ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3074624/]
- 35. Reliability and accuracy of single-molecule FRET studies ... [https://www.nature.com/articles/s41592-023-01807-0]
- 36. Robust calibration and quantification of FRET signals using ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12595091/]
- 37. Analytical ultracentrifugation as a tool for studying protein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3652485/]
- 38. Analytical ultracentrifugation for the study of protein ... [https://www.sciencedirect.com/science/article/abs/pii/S1367593106001244]
- 39. Characterization of STAT self-association by analytical ultracentrifugation - PubMed [https://pubmed.ncbi.nlm.nih.gov/23296732/]
- 40. A High-Throughput Fluorescence Polarization-Based Assay for the SH2 Domain of STAT4 - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9781101/]
- 41. Evolving cognition of the JAK-STAT signaling pathway [https://www.nature.com/articles/s41392-023-01468-7]
- 42. Novel STAT3 Inhibitors Targeting STAT3 Dimerization by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9196127/]
- 43. Comparison of a molecular dynamics model with the X-ray ... [https://pubmed.ncbi.nlm.nih.gov/21724649/]
- 44. STAT5B leukemic mutations, altering SH2 tyrosine 665 ... [https://www.life-science-alliance.org/content/8/7/e202503222]
- 45. Disease-Associated Mutations of the STAT5B SH2 Domain ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11958444/]
- 46. Deep mutational scanning of the multi-domain ... [https://www.nature.com/articles/s41467-025-60641-4]
- 47. The SH2 Domain Interaction Landscape [https://www.sciencedirect.com/science/article/pii/S2211124713001083]
- 48. The SH2 domain interaction landscape - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC4110347/]
- 49. STAT proteins: a kaleidoscope of canonical and non ... [https://jhoonline.biomedcentral.com/articles/10.1186/s13045-021-01214-y]
- 50. Roles of unphosphorylated STATs in signaling [https://pubmed.ncbi.nlm.nih.gov/18364677/]
- 51. Mutational Analysis of the STAT6 SH2 Domain [https://www.sciencedirect.com/science/article/pii/S002192581880535X]
- 52. CDD Conserved Protein Domain Family: SH2_STAT1 [https://www.ncbi.nlm.nih.gov/Structure/cdd/cd10372]
- 53. Structure of the Unphosphorylated STAT5a Dimer [https://www.sciencedirect.com/science/article/pii/S0021925820590455]
- 54. Canonical and non-canonical JAK–STAT signaling - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3082280/]
- 55. Biomolecular Condensation of SH Domain-Containing... | SpringerLink 2 [https://link.springer.com/protocol/10.1007/978-1-0716-3393-9_20]
- 56. Liquid–liquid phase separation in human health and ... [https://www.nature.com/articles/s41392-021-00678-1]
- 57. Liquid–Liquid Phase Separation in Cancer Signaling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8997759/]
- 58. - Liquid in innate immunity - PMC liquid phase separation [https://pmc.ncbi.nlm.nih.gov/articles/PMC11247960/]
- 59. The genetics of hyper IgE syndromes - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC11876172/]
- 60. An update on the hyper-IgE syndromes - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC3674633/]
- 61. STAT3-confusion-of-function: Beyond the loss and gain ... [https://www.sciencedirect.com/science/article/abs/pii/S0091674922008375]
- 62. SH2-domain mutations in STAT3 in hyper-IgE syndrome ... [https://pubmed.ncbi.nlm.nih.gov/21792878/]
- 63. JAK/STAT pathway directed therapy of T-cell leukemia ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5469693/]
- 64. Oncogenic activation of JAK3-STAT signaling confers ... [https://www.nature.com/articles/s41375-017-0004-x]
- 65. Role of the STAT1-SH2 Domain and STAT2 in the ... [https://www.sciencedirect.com/science/article/pii/S0021925819592043]
- 66. A reinterpretation of the dimerization interface of the N- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2312425/]
- 67. A mutational hotspot that determines highly repeatable ... [https://www.nature.com/articles/s41467-021-26286-9]
- 68. Malignant JAK-signaling: at the interface of inflammation ... [https://www.nature.com/articles/s41375-025-02569-8]
- 69. Allosteric regulation in STAT 3 interdomains is mediated by a rigid core... [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1010794]
- 70. A Mutation in the SH2 Domain of STAT2 Prolongs Tyrosine ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1924825/]
- 71. STAT5B leukemic mutations, altering SH2 tyrosine 665, have ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11999048/]
- 72. The role of JAK- STAT and its regulators in the fate... signaling pathway [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-017-0177-y]
- 73. Alternative ways of modulating JAK-STAT pathway [https://pmc.ncbi.nlm.nih.gov/articles/PMC3670285/]
- 74. Post-translational modifications of Stat3: The state of the art [https://www.sciencedirect.com/science/article/abs/pii/S0898656825004632]
- 75. Potential signaling pathways, biomarkers, natural drugs ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1615770/full]
- 76. Molecular basis of IFN-γ–induced STAT3 phosphorylation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12552534/]
- 77. Update on Structure and Function of SH2 Domains [https://pubmed.ncbi.nlm.nih.gov/41009621/]
- 78. Recludix Unveils First BTK SH2 Domain Inhibitor [https://www.dermatologytimes.com/view/recludix-unveils-first-btk-sh2-domain-inhibitor]
- 79. Disease-Associated Mutations of the STAT5B SH2 Domain ... [https://link.springer.com/article/10.1007/s10911-025-09582-8]
- 80. Development of a Peptide Inhibitor Targeting the C-SH2 ... [https://pubmed.ncbi.nlm.nih.gov/40318117/]
- 81. Decoding the selective mechanism behind a monobody ... [https://pubs.rsc.org/en/content/articlelanding/2025/cp/d5cp00211g]
- 82. JAK-STAT信号通路:机制、功能与靶向治疗全景解读 [https://www.cn-healthcare.com/articlewm/20251113/content-1660716.html]
- 83. Advances of the Correlation between JAK-STAT3 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6000523/]
- 84. 导致高IgE综合征的STAT3突变小鼠模型与人类病理学不同。 [https://suppr.wilddata.cn/browses/detail/pubmed/40868996]
- 85. Activating mutations of STAT5B and STAT3 in lymphomas ... [https://www.nature.com/articles/ncomms7025]
- 86. STAT3 and STAT5B Mutations in T/NK-Cell Chronic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7760806/]
- 87. CN114269763A - Stat3的小分子降解剂 [https://patents.google.com/patent/CN114269763A/zh]
- 88. A family-wide assessment of latent STAT transcription factor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10200994/]
- 89. Structural characterization of unphosphorylated STAT5a ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2762584/]
- 90. Structural Bases of Unphosphorylated STAT1 Association ... [https://www.sciencedirect.com/science/article/pii/S1097276505011202]
- 91. STAT heterodimers in immunity: A mixed message or ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3670269/]
- 92. Specificity of signaling by STAT1 depends on SH2 and C ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC140204/]
- 93. Systematic characterization of the specificity of the SH2 ... [https://www.sciencedirect.com/science/article/abs/pii/S1874391913000080]
- 94. Binding Specificity of SH2 Domains: Insight from Free ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2635922/]
- 95. protein heterodimer structure prediction by template-based ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8262733/]
- 96. Structure-guided development of heterodimer-selective ... [https://www.nature.com/articles/ncomms12298]
- 97. Allosteric inhibitors of the STAT3 signaling pathway [https://www.sciencedirect.com/science/article/abs/pii/S0223523420300891]
- 98. Selective inhibition of STAT3 signaling using monobodies ... [https://www.nature.com/articles/s41467-020-17920-z]
- 99. Allosteric Communication Across STAT3 Domains ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4767637/]
- 100. An allosteric inhibitor targeting the STAT3 coiled- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11969721/]
- 101. Structure-Guided Expansion Strategy Unveils Potent ... [https://www.sciencedirect.com/science/article/abs/pii/S0223523425007536]
- 102. Targeting SHP2 with Natural Products: Exploring Saponin ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12109979/]