Optimizing MgCl2 Concentration to Eliminate Nonspecific PCR Bands: A Strategic Guide for Researchers
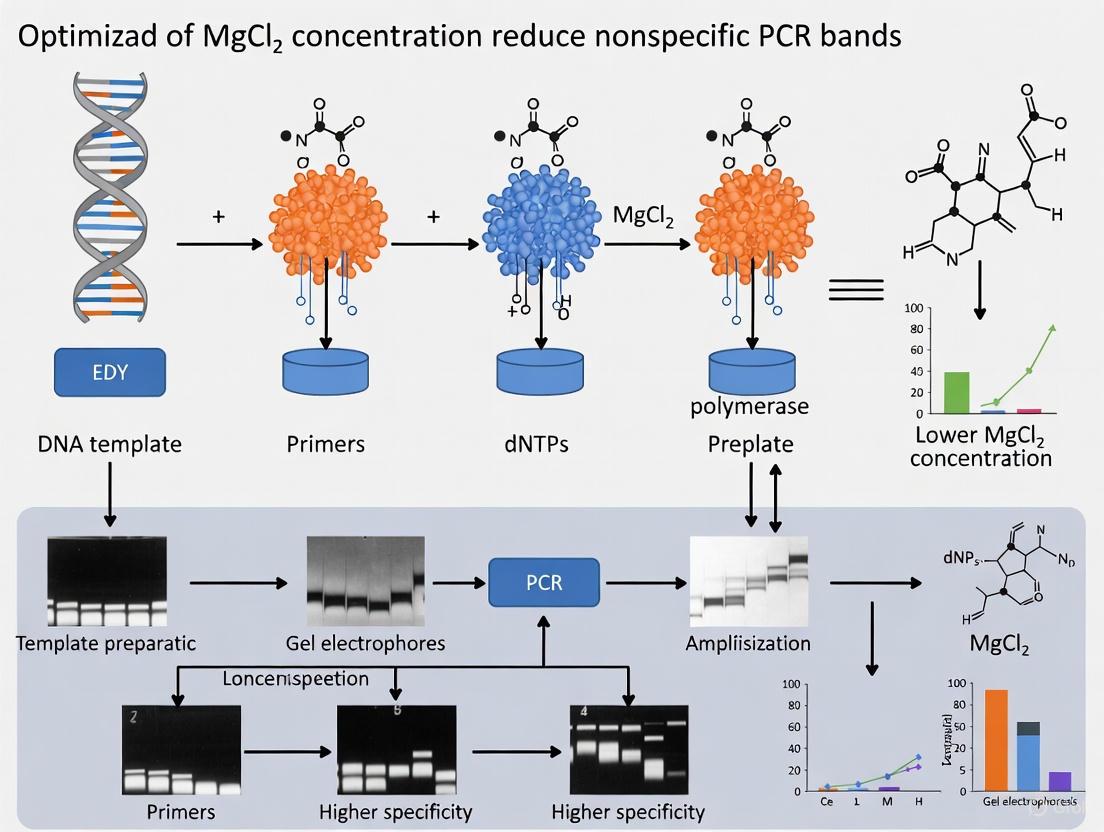
Nonspecific amplification and multiple bands on agarose gels are common, time-consuming challenges in PCR, often stemming from suboptimal magnesium chloride (MgCl2) concentration.
Optimizing MgCl2 Concentration to Eliminate Nonspecific PCR Bands: A Strategic Guide for Researchers
Abstract
Nonspecific amplification and multiple bands on agarose gels are common, time-consuming challenges in PCR, often stemming from suboptimal magnesium chloride (MgCl2) concentration. This article provides a comprehensive, evidence-based guide for researchers and drug development professionals to systematically optimize MgCl2 to enhance PCR specificity and efficiency. We cover the foundational role of Mg2+ as a DNA polymerase cofactor, present step-by-step methodological optimization and troubleshooting protocols, and review advanced validation techniques and predictive modeling. By synthesizing current research and quantitative data, this resource aims to equip scientists with the knowledge to reliably produce clean, specific amplification for critical applications in biomedical and clinical research.
The Critical Role of MgCl2 in PCR: Understanding the Science Behind the Co-factor
MgCl2 as an Essential Cofactor for DNA Polymerase Activity
Magnesium chloride (MgCl₂) is an indispensable component of the polymerase chain reaction (PCR), functioning as a critical cofactor for DNA polymerase enzyme activity. Without Mg²⁺ ions, DNA polymerases remain enzymatically inactive, unable to catalyze the replication of DNA templates [1]. This ion serves dual essential roles: it enhances the catalytic function of the DNA polymerase enzyme and facilitates the specific binding of primers to their target DNA sequences [1] [2]. The precise optimization of MgCl₂ concentration is fundamental to successful PCR amplification, directly influencing reaction efficiency, specificity, and yield, while insufficient or excessive amounts can lead to amplification failure or nonspecific products [1].
Mechanisms of Action: How MgCl₂ Functions in PCR
Molecular Mechanism for DNA Polymerase Activation
The magnesium ion (Mg²⁺) derived from MgCl₂ is fundamental to the catalytic mechanism of DNA synthesis. During the extension phase of PCR, Mg²⁺ ions bind directly to deoxynucleoside triphosphates (dNTPs) at their alpha phosphate groups [1] [2]. This binding facilitates the removal of beta and gamma phosphates, enabling the resulting deoxynucleoside monophosphate (dNMP) to form a phosphodiester bond with the 3' hydroxyl group (3'-OH) of the adjacent nucleotide on the growing DNA strand [1]. This catalytic process occurs at the active site of DNA polymerase, where Mg²⁺ serves as a essential bridge for the nucleotidyl transferase reaction [3].
Mechanism for Primer-Template Binding
MgCl₂ significantly influences the hybridization dynamics between primers and template DNA by modulating electrostatic interactions. The magnesium cations bind to the negatively charged phosphate groups along the DNA backbone, effectively neutralizing the natural electrostatic repulsion that occurs between two complementary DNA strands [1] [2]. This stabilization promotes proper annealing of primers to their specific target sequences and increases the melting temperature (Tm) of the DNA duplex [1]. Research indicates that every 0.5 mM increase in MgCl₂ concentration raises the DNA melting temperature by approximately 1.2°C within the optimal concentration range [4].
Figure 1: Dual mechanistic roles of Mg²⁺ in PCR. Mg²⁺ ions activate DNA polymerase, facilitate dNTP incorporation, and stabilize primer-template binding.
Optimization Guidelines: MgCl₂ Concentration Effects
Quantitative Effects on PCR Performance
The concentration of MgCl₂ profoundly impacts multiple aspects of PCR performance, with specific quantitative relationships observed between Mg²⁺ concentration and reaction outcomes:
Table 1: Quantitative Effects of MgCl₂ Concentration on PCR Parameters
| Parameter | Effect of Low MgCl₂ (<1.5 mM) | Effect of Optimal MgCl₂ (1.5-3.0 mM) | Effect of High MgCl₂ (>3.0 mM) |
|---|---|---|---|
| DNA Polymerase Activity | Significantly reduced catalytic efficiency; insufficient cofactor availability [1] | Maximum enzymatic activity; optimal dNTP incorporation rates [1] [4] | Saturated enzyme activity; potential inhibition or error-prone synthesis [1] |
| Primer Annealing Specificity | Reduced primer-template stability; weak or non-existent binding [1] | Specific binding to target sequences with appropriate duplex stability [1] | Non-specific primer binding; mismatched annealing [1] [5] |
| Amplification Yield | Minimal or no product formation; weak amplification [1] | High yield of desired specific product [4] | Multiple non-specific products; primer-dimer formation [1] [5] |
| Melting Temperature (Tₘ) | Decreased DNA duplex stability [1] | Optimal Tₘ with 1.2°C increase per 0.5 mM MgCl₂ [4] | Excessive duplex stability; impaired denaturation [1] |
Template-Specific Concentration Requirements
Different template characteristics necessitate adjustment of MgCl₂ concentrations beyond standard protocols. Evidence from meta-analyses indicates that template complexity significantly influences optimal Mg²⁺ requirements, with genomic DNA templates typically requiring higher concentrations than simpler templates such as plasmid DNA or cDNA [4]. GC-rich templates often benefit from slightly elevated MgCl₂ concentrations (2.0-3.0 mM) to overcome the increased stability of GC base pairs [6]. Additionally, the presence of PCR inhibitors in DNA extracts may necessitate increased MgCl₂ concentrations, as these compounds can sequester available Mg²⁺ ions and reduce their effective concentration in the reaction [1].
Table 2: Recommended MgCl₂ Concentration Ranges by Template Type
| Template Type | Recommended MgCl₂ Range | Special Considerations |
|---|---|---|
| Standard Templates | 1.5 - 2.0 mM [5] [4] | Suitable for most applications with typical GC content (40-60%) |
| Genomic DNA | 2.0 - 3.0 mM [4] | Higher complexity requires increased Mg²⁺ for efficient amplification |
| GC-Rich Sequences | 1.5 - 2.5 mM [6] | Enhanced stability of GC bonds may require optimization within this range |
| Plasmid DNA | 1.0 - 1.5 mM [3] | Lower complexity enables reduced Mg²⁺ requirements |
| Inhibitor-Containing Samples | 2.5 - 4.0 mM [1] | Increased concentration compensates for Mg²⁺ binding by inhibitors |
Troubleshooting Guide: MgCl₂-Related PCR Issues
Frequently Asked Questions
Q: What specific problems occur with excessive MgCl₂ in PCR? A: Elevated MgCl₂ concentrations (typically >3.0 mM) promote non-specific primer binding, resulting in multiple erroneous amplification products visible as extraneous bands on agarose gels [1] [5]. Excessive Mg²⁺ also increases the likelihood of primer-dimer formation due to stabilized non-productive primer interactions [5]. These issues manifest electrophoretically as a ladder or smear of DNA fragments rather than a single discrete band at the expected amplicon size [1] [7].
Q: How does insufficient MgCl₂ affect PCR outcomes? A: Inadequate MgCl₂ (<1.5 mM) causes dramatic reductions in amplification efficiency, resulting in weak product yield or complete PCR failure [1]. This occurs because DNA polymerase activity is strictly dependent on Mg²⁺ cofactors; without sufficient magnesium, the enzyme cannot catalyze DNA strand elongation effectively [1] [3]. Primer-template binding is also compromised under low Mg²⁺ conditions due to insufficient stabilization of the DNA duplex [1].
Q: What is the recommended approach for optimizing MgCl₂ concentration? A: Implement a titration experiment testing MgCl₂ concentrations across a range of 1.0-4.0 mM in 0.5 mM increments [4] [8]. The optimal concentration produces a single strong band of the expected size with minimal background or non-specific products [1]. For challenging templates (GC-rich, genomic DNA, or inhibitor-containing samples), extend the titration range upward to 4.5 mM while monitoring for specificity loss [5] [4].
Q: How does MgCl₂ concentration interact with PCR additives? A: MgCl₂ concentration should be re-optimized when introducing PCR enhancers such as DMSO, betaine, or formamide [8] [6]. These additives alter DNA duplex stability and primer annealing dynamics, effectively changing Mg²⁺ requirements [6]. For example, when using DMSO for GC-rich templates, optimal MgCl₂ concentrations typically range between 1.5-2.0 mM rather than standard concentrations [6].
Diagnostic Flowchart for MgCl₂ Optimization
Figure 2: Systematic troubleshooting approach for MgCl₂-related PCR issues. This flowchart guides optimization based on specific amplification problems.
Experimental Protocols: MgCl₂ Optimization Methods
Standard MgCl₂ Titration Protocol
Objective: Determine the optimal MgCl₂ concentration for specific PCR amplification [4] [8].
Reagents and Equipment:
- 10X PCR Buffer (without MgCl₂)
- 25 mM MgCl₂ stock solution
- DNA template (e.g., genomic DNA, plasmid)
- Forward and reverse primers (20 μM each)
- dNTP mix (10 mM total)
- DNA polymerase (e.g., Taq polymerase)
- Sterile distilled water
- Thermal cycler
- Agarose gel electrophoresis system
Procedure:
- Prepare a master mixture containing all PCR components except MgCl₂ and DNA template [8].
- Aliquot the master mixture into 8 PCR tubes (0.2 mL thin-walled).
- Add MgCl₂ stock solution to achieve these final concentrations: 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, and 4.5 mM [4].
- Add DNA template to each tube and mix gently by pipetting.
- Perform PCR amplification using appropriate cycling parameters:
- Analyze PCR products by agarose gel electrophoresis (1.5-2.0% agarose).
- Identify the MgCl₂ concentration that produces a single, intense band of the expected size with minimal background [1] [4].
Specialized Protocol for GC-Rich Templates
Objective: Amplify challenging GC-rich DNA sequences through combined MgCl₂ and additive optimization [6].
Modified Procedure:
- Prepare MgCl₂ titration series as in the standard protocol (1.0-4.0 mM range).
- Include PCR enhancers in the reaction mixture:
- Increase annealing temperature 5-10°C above the calculated primer Tₘ to enhance specificity [6].
- Extend denaturation time to 1-2 minutes to ensure complete separation of GC-rich strands.
- Implement a "touchdown" PCR protocol if standard optimization fails, gradually decreasing annealing temperature over initial cycles [7].
Expected Results: For the EGFR promoter sequence (GC-rich), successful amplification typically requires 1.5-2.0 mM MgCl₂ combined with 5% DMSO and elevated annealing temperatures [6].
Research Reagent Solutions
Table 3: Essential Reagents for MgCl₂ Optimization Experiments
| Reagent/Category | Specific Function in PCR | Optimization Considerations |
|---|---|---|
| MgCl₂ Stock Solution | Primary source of Mg²⁺ cofactors for DNA polymerase activity and primer-template stabilization [1] | Use high-purity, molecular biology grade; prepare fresh solutions to prevent oxidation; typical stock concentration is 25 mM [8] |
| PCR Buffer Systems | Provides optimal pH and ionic environment; may contain supplemental MgCl₂ [8] | Verify Mg²⁺ content in commercial buffers; use Mg²⁺-free buffers for precise optimization experiments [8] [3] |
| DNA Polymerase | Enzyme that catalyzes DNA strand elongation using dNTPs and Mg²⁺ cofactors [1] [3] | Taq polymerase most common; hot-start variants reduce non-specific amplification; 1-2 units per 50 μL reaction [8] [3] |
| PCR Enhancers | Modifies nucleic acid melting behavior to facilitate amplification of challenging templates [8] [6] | DMSO (1-10%), betaine (0.5-2.5 M), or formamide (1.25-10%); requires re-optimization of MgCl₂ concentration [8] [6] |
| dNTP Mix | Building blocks for DNA synthesis; substrates for DNA polymerase activity [3] | Standard concentration 200 μM each dNTP; Mg²⁺ binds dNTPs, reducing free Mg²⁺ availability - adjust accordingly [1] [3] |
In polymerase chain reaction (PCR), the magnesium ion (Mg2+) is an essential cofactor that directly influences the efficiency and specificity of DNA amplification. Within the context of troubleshooting nonspecific PCR bands, understanding Mg2+'s dual role is paramount. It facilitates the binding of primers to their template DNA and is a critical component for the catalytic activity of DNA polymerase. An imbalance in Mg2+ concentration is a frequent cause of nonspecific amplification, leading to multiple spurious bands on an agarose gel. This guide details the molecular mechanisms of Mg2+ and provides a systematic, evidence-based approach to optimizing MgCl2 concentration to achieve clean, specific PCR results.
The Molecular Role of Mg2+ in PCR
Magnesium ions are fundamental to the PCR process at a molecular level, primarily functioning in two key areas: enzyme catalysis and nucleic acid stabilization.
Mg2+ as a Catalytic Cofactor
The DNA polymerase enzyme requires Mg2+ for its catalytic activity. The ion plays a direct role in the chemical reaction of DNA synthesis. It facilitates the formation of a phosphodiester bond between the 3'-hydroxyl (3'-OH) group of the primer and the phosphate group of an incoming deoxynucleoside triphosphate (dNTP) [3]. Specifically, Mg2+ binds to the dNTP at its α-phosphate group, which allows for the removal of the β and gamma phosphates and helps catalyze the bond formation [9]. Without Mg2+ present at the active site of the polymerase, the incorporation of nucleotides cannot proceed efficiently.
Mg2+ in Electrostatic Stabilization and Primer Binding
Beyond the active site, Mg2+ is crucial for stabilizing the overall structure of the nucleic acid complex. The backbone of DNA is highly negatively charged due to its phosphate groups. This creates electrostatic repulsion between the primer and the template DNA strand, hindering efficient binding. Mg2+ ions, with their positive charge, act as shields that neutralize this repulsion [3] [9]. By binding to the phosphate backbones, Mg2+ reduces the electrostatic barrier, allowing the primer to anneal to its complementary sequence on the template DNA with greater stability and specificity. This function is critically important for reducing mis-priming, where primers bind to partially complementary, off-target sites, a primary source of nonspecific amplification [10] [3].
Troubleshooting Guide: Resolving Nonspecific PCR Bands via Mg2+ Optimization
Nonspecific PCR products, visible as multiple or smeared bands on a gel, are a common issue often linked to suboptimal Mg2+ concentrations. The following questions and answers provide a targeted troubleshooting guide.
FAQ 1: Why does high Mg2+ concentration cause nonspecific bands in my PCR?
Excess Mg2+ in a PCR reaction is a frequent cause of nonspecific amplification for two key reasons:
- Reduced Primer Stringency: High concentrations of Mg2+ excessively stabilize the DNA duplex, even imperfect matches. This reduces the reaction's stringency, allowing primers to anneal to non-target DNA sequences that have partial complementarity [11] [10].
- Stimulation of Non-Specific Enzyme Activity: Elevated Mg2+ levels can enhance the general activity of the DNA polymerase, making it more likely to extend these mis-annealed primers, leading to a ladder or smear of incorrect products [11].
Recommendation: If you observe multiple bands, the first step is to lower the Mg2+ concentration incrementally. A gradient PCR is highly recommended to find the optimal concentration for your specific primer-template system [11] [10].
FAQ 2: I have no PCR product. Could Mg2+ be the problem?
Yes. While excess Mg2+ causes nonspecificity, insufficient Mg2+ can lead to a complete failure of amplification or very low yield. This is because:
- Impaired Polymerase Function: The DNA polymerase enzyme strictly requires Mg2+ as a cofactor. At very low concentrations, the enzyme's activity is drastically reduced, as it cannot efficiently catalyze the incorporation of dNTPs [9].
- Unstable Primer-Template Complexes: Inadequate Mg2+ fails to sufficiently neutralize the electrostatic repulsion between the primer and template. This results in unstable hybrids that dissociate before the polymerase can initiate synthesis [3].
Recommendation: If there is no product, try increasing the Mg2+ concentration in small steps (e.g., 0.5 mM increments) to restore polymerase activity and stabilize primer binding [9].
FAQ 3: How do I systematically optimize Mg2+ concentration to eliminate nonspecific bands?
A methodical approach is required to pinpoint the ideal Mg2+ concentration. The following protocol provides a detailed methodology.
Experimental Protocol: Optimizing MgCl2 Concentration
Objective: To determine the MgCl2 concentration that yields maximum specific amplification of the target DNA fragment with minimal to no nonspecific background.
Materials and Reagents:
- DNA template (e.g., genomic DNA, plasmid)
- Forward and reverse primers
- 10X PCR buffer (without MgCl2)
- MgCl2 solution (e.g., 25 mM or 50 mM)
- dNTP mix (10 mM)
- DNA polymerase (e.g., Taq)
- Nuclease-free water
- PCR tubes and thermal cycler
Procedure:
- Prepare a Master Mix: Create a master mix for n+1 reactions (where 'n' is the number of Mg2+ conditions to test) containing all common components: water, 1X PCR buffer, primers (0.1–1 µM each), dNTPs (200 µM each), DNA template (e.g., 5–50 ng gDNA), and DNA polymerase (0.5–2.5 units/50 µL reaction) [8].
- Aliquot the Master Mix: Dispense equal volumes of the master mix into a series of PCR tubes.
- Add MgCl2: Add MgCl2 to each tube to create a concentration gradient. A typical range is 0.5 mM to 4.0 mM, in 0.5 mM increments [9].
- Run PCR: Place the tubes in a thermal cycler and run the optimized cycling program for your target.
- Analyze Results: Separate the PCR products by agarose gel electrophoresis. Identify the MgCl2 concentration that produces a single, intense band of the expected size with the cleanest background.
Quantitative Mg2+ Optimization Data
The table below summarizes the effects of varying MgCl2 concentrations and provides recommended starting points for optimization.
Table 1: Effects of MgCl2 Concentration on PCR Outcomes and Optimization Strategies
| MgCl2 Concentration | Observed Outcome on Gel | Molecular Cause | Recommended Action |
|---|---|---|---|
| Too Low (< 1.0 mM) | No product, or very faint target band [9] | Insufficient polymerase cofactor activity; unstable primer-template complexes [3] | Increase concentration in 0.5 mM increments [9] |
| Optimal (1.5 - 2.5 mM)* | Single, bright band of correct size | Balanced catalysis and primer binding stringency | Maintain this concentration for future experiments |
| Too High (> 3.0 mM) | Multiple bands, smearing, or high background [11] [10] | Reduced primer annealing stringency; promotion of non-specific extension [9] | Decrease concentration in 0.5 mM increments [11] |
*Note: The optimal range is a common starting point but can vary based on polymerase, buffer composition, and template. A gradient test is essential [8].
The Scientist's Toolkit: Essential Reagents for PCR Optimization
Successful PCR troubleshooting relies on high-quality reagents. The table below lists key materials and their functions.
Table 2: Key Research Reagent Solutions for PCR Optimization
| Reagent | Function in PCR | Key Considerations for Optimization |
|---|---|---|
| DNA Polymerase | Enzymatically synthesizes new DNA strands. | Use hot-start polymerases to prevent non-specific priming at low temperatures [11] [10]. For GC-rich targets, use specialized polymerases with GC enhancers [9]. |
| MgCl2 Solution | Essential cofactor for polymerase; stabilizes primer-template binding. | The only component that requires extensive concentration titration (typically 0.5-4.0 mM) for each new primer set [9] [8]. |
| PCR Additives | Modifies DNA melting behavior and improves reaction specificity/yield. | DMSO, Betaine, or Glycerol can help denature GC-rich secondary structures. Formamide can increase primer stringency [9]. |
| High-Purity dNTPs | Building blocks for new DNA strands. | Use equimolar concentrations to maintain polymerase fidelity. Excess dNTPs can chelate Mg2+, effectively reducing its free concentration [11] [3]. |
Integrated Optimization Workflow
The following diagram illustrates the logical decision-making process for troubleshooting nonspecific PCR bands, with a central focus on Mg2+ optimization.
Achieving specific amplification in PCR is a cornerstone of reliable molecular biology data. As detailed in this guide, the concentration of Mg2+ is a pivotal factor controlling the fine balance between primer binding stability, enzymatic catalysis, and reaction stringency. A systematic approach, beginning with a Mg2+ gradient titration and incorporating secondary checks of annealing temperature and reagent quality, provides a robust pathway to eliminating nonspecific bands. Mastery of Mg2+ optimization empowers researchers to significantly enhance the reproducibility and specificity of their PCR experiments, thereby supporting the generation of high-quality data for scientific and diagnostic applications.
Troubleshooting Guides
FAQ: How does MgCl₂ concentration specifically affect my PCR results?
MgCl₂ is a critical cofactor for DNA polymerase activity, and its concentration directly influences both the enzyme's efficiency and its accuracy in primer binding. An imbalance often manifests in two ways:
- Too much MgCl₂ (typically >3.0-4.0 mM) reduces the stringency of primer annealing. This allows primers to bind to non-target sites with partial sequence similarity, resulting in non-specific amplification. You will observe this as multiple bands or a smeared background on an agarose gel [12] [13].
- Too little MgCl₂ (typically <1.5 mM) compromises DNA polymerase activity and stabilizes the DNA duplex less effectively. This leads to greatly reduced yield or complete PCR failure due to inefficient primer extension and poor enzyme function [12] [14].
FAQ: What is the recommended starting point and optimal range for MgCl₂?
For most standard PCR applications, a final MgCl₂ concentration of 1.5 mM is a common and safe starting point [12]. However, extensive research has identified an optimal functional range.
Table 1: Evidence-Based MgCl₂ Concentration Guidelines [15] [4]
| Parameter | Recommended Range | Key Quantitative Finding |
|---|---|---|
| Overall Optimal Range | 1.5 - 3.0 mM | Synthesized from a meta-analysis of 61 studies. |
| Effect on DNA Melting Temperature (Tₘ) | --- | Every 0.5 mM increase in MgCl₂ raises DNA Tₘ by ~1.2°C within the 1.5-3.0 mM range. |
| Template-Specific Adjustment | Higher end for complex templates (e.g., genomic DNA) | Genomic DNA templates generally require higher MgCl₂ concentrations than simple plasmid DNA. |
Troubleshooting Table: Diagnosing and Solving MgCl₂-Related PCR Problems
Table 2: Troubleshooting Guide for MgCl₂ Optimization
| Observed Result | Potential Cause | Recommended Solution |
|---|---|---|
| Multiple non-specific bands or smearing on gel [12] [13] | MgCl₂ concentration too high, reducing annealing stringency. | Create a MgCl₂ gradient (e.g., 1.0, 1.5, 2.0, 2.5, 3.0 mM) and re-run the PCR. Decrease concentration in 0.5 mM increments. |
| No product or very faint band on gel [12] | MgCl₂ concentration too low for polymerase activity and primer binding. | Perform a MgCl₂ gradient as above, but focus on increasing the concentration in 0.5 mM steps up to 4.0 mM. |
| PCR failure with GC-rich templates (>60% GC) [12] | Standard MgCl₂ conditions cannot overcome stable secondary structures. | Combine optimization strategies: Use a specialized polymerase, include additives like DMSO or betaine, and titrate MgCl₂ (often requiring higher concentrations). |
Experimental Protocols
Step-by-Step Protocol: MgCl₂ Titration for PCR Optimization
This protocol provides a detailed methodology for empirically determining the ideal MgCl₂ concentration for any new PCR assay.
Principle: By setting up a series of identical reactions that vary only in MgCl₂ concentration, you can directly visualize which condition provides the strongest specific amplification with the least background.
Materials:
- 10X PCR Buffer (without MgCl₂): Provides the core reaction environment (e.g., Tris-HCl, KCl) [8].
- MgCl₂ Stock Solution (25 mM): The variable component for titration.
- dNTP Mix (10 mM): Building blocks for DNA synthesis [8] [14].
- Forward and Reverse Primers (20 μM each): Designed for your specific target [8].
- DNA Polymerase (e.g., Taq, 5 U/μL): The enzyme catalyzing the reaction [14].
- Template DNA: The DNA you wish to amplify [14].
- Nuclease-Free Water: To adjust the final volume.
Procedure:
- Prepare a Master Mix: Combine all common reagents in a single tube to minimize pipetting errors and ensure consistency across tubes. Calculate for ( n+1 ) reactions, where ( n ) is the number of MgCl₂ conditions you are testing.
Component Volume per 50 μL Reaction Final Concentration 10X PCR Buffer (no MgCl₂) 5 μL 1X dNTP Mix (10 mM) 1 μL 200 μM each Forward Primer (20 μM) 1 μL 0.4 μM Reverse Primer (20 μM) 1 μL 0.4 μM DNA Polymerase (5 U/μL) 0.5 μL 2.5 U Template DNA X μL (e.g., 1-100 ng) Variable Nuclease-Free Water To 49.5 μL (before MgCl₂) ---
Aliquot the Master Mix: Pipette 49.5 μL of the Master Mix into each PCR tube.
Add MgCl₂: Add the 25 mM MgCl₂ stock solution to each tube to create your desired concentration gradient. Table 3: Example Setup for a MgCl₂ Titration Experiment
Tube Volume of 25 mM MgCl₂ Stock (μL) Final MgCl₂ Concentration (mM) 1 1.0 1.0 2 1.5 1.5 3 2.0 2.0 4 2.5 2.5 5 3.0 3.0 6 4.0 4.0 Run PCR: Place the tubes in a thermal cycler and start the optimized cycling program.
Analyze Results: Separate the PCR products by agarose gel electrophoresis. Identify the tube with the most intense specific band and the cleanest background.
Workflow Diagram: MgCl₂ Optimization Logic
The following diagram outlines the logical decision-making process for troubleshooting and optimizing MgCl₂ in your PCR experiments.
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Reagents for PCR and MgCl₂ Optimization
| Reagent / Material | Critical Function in PCR | Role in MgCl₂ Optimization |
|---|---|---|
| MgCl₂ Stock Solution | Serves as a cofactor for DNA polymerase; stabilizes primer-template binding and negatively charged dNTPs [14]. | The primary variable in the optimization experiment. A pure, high-quality stock is essential. |
| PCR Buffer (without MgCl₂) | Provides the optimal ionic environment (pH, salt) for polymerase activity and DNA stability. | Using a MgCl₂-free buffer is crucial for a true titration, as it gives you full control over the Mg²⁺ concentration. |
| DNA Polymerase | Enzyme that synthesizes new DNA strands by adding dNTPs to the primer. | Polymerase activity is directly dependent on Mg²⁺. Specialized polymerases for GC-rich targets may have different optimal Mg²⁺ ranges [12]. |
| dNTP Mix | The four deoxynucleotides (dATP, dCTP, dGTP, dTTP) are the building blocks for DNA synthesis. | dNTPs chelate Mg²⁺ ions. The standard 0.2 mM dNTP concentration must be considered, as it affects the amount of free Mg²⁺ available for the polymerase [14]. |
| PCR Additives (DMSO, Betaine) | Assist in denaturing difficult templates (e.g., GC-rich DNA) by reducing secondary structure formation [12] [8]. | When using these additives, the optimal MgCl₂ concentration may shift, requiring re-optimization of the Mg²⁺ balance in the new reaction environment. |
In polymerase chain reaction (PCR) optimization, controlling nonspecific amplification is a fundamental challenge that can compromise experimental results. The concentration of magnesium chloride (MgCl₂) is a critical factor in this process, directly influencing reaction efficiency and specificity through a defined logarithmic relationship with DNA melting temperature (Tₘ). This guide provides researchers with targeted troubleshooting and quantitative protocols to harness this relationship, enabling precise MgCl₂ optimization to eliminate smeared bands and enhance PCR fidelity.
FAQs: Troubleshooting MgCl₂ in PCR
1. How does MgCl₂ concentration specifically lead to smeared or nonspecific PCR bands?
Smeared bands on an agarose gel indicate the presence of nonspecific PCR products or DNA fragments of varying sizes. Suboptimal MgCl₂ concentration is a primary cause. Excessive MgCl₂ reduces the reaction stringency, facilitating primer binding to incorrect, off-target sites on the DNA template. This promotes nonspecific amplification and smearing [11] [16]. Conversely, insufficient MgCl₂ can critically impair DNA polymerase activity, potentially leading to weak or no amplification [10].
2. What is the quantitative relationship between MgCl₂ and DNA melting temperature?
A comprehensive meta-analysis of peer-reviewed studies established a clear logarithmic relationship between MgCl₂ concentration and DNA melting temperature. Within the optimal concentration range of 1.5 to 3.0 mM, every 0.5 mM increase in MgCl₂ is associated with an average increase of 1.2 °C in the DNA melting temperature [15]. This quantitative relationship is foundational for predicting and controlling primer annealing efficiency.
3. What is the recommended MgCl₂ concentration range for optimizing a standard PCR?
While the optimal range must be determined empirically for each primer-template system, general guidelines exist. A typical MgCl₂ titration should be performed within a range of 1.5 mM to 5.0 mM [16]. For most standard PCRs, the final MgCl₂ concentration falls between 1.5 and 3.0 mM [15]. It is crucial to adjust this based on template complexity; genomic DNA often requires higher concentrations than simple plasmid DNA [15].
4. What are the consequences of using a MgCl₂ concentration that is too high or too low?
The balance is critical, as both high and low concentrations cause distinct problems:
- Too High (>3.0-5.0 mM): Promotes nonspecific binding and smeared bands, reduces fidelity by increasing misincorporation of nucleotides [11].
- Too Low (<1.5 mM): Results in significantly reduced or failed amplification due to inefficient primer annealing and impaired DNA polymerase activity [10].
5. How do I systematically troubleshoot a PCR experiment producing smeared bands?
Begin by addressing the most common causes related to MgCl₂ and reaction conditions:
- Optimize MgCl₂: Perform a titration experiment in 0.5 mM increments [16].
- Check Template Quantity: Excess DNA template is a common cause of smearing; try serial dilutions [17] [16].
- Increase Annealing Temperature: A higher temperature enhances specificity. Use the established MgCl₂-Tₘ relationship as a guide [17] [15].
- Use Hot-Start Polymerase: This prevents nonspecific amplification during reaction setup [11].
- Ensure Reagent Purity: Use fresh aliquots to avoid contaminants that can cause degradation and smearing [17].
Quantitative Data and Protocols
MgCl₂ Titration Guide
The following table provides a standard setup for a 50 μL PCR reaction to empirically determine the optimal MgCl₂ concentration. A negative control (without DNA template) should be included to check for contamination.
Table 1: Experimental setup for MgCl₂ titration in a 50 µL PCR reaction.
| Reagent | Initial Concentration | Master Mix (for 1 rxn) | Final Concentration |
|---|---|---|---|
| Sterile Water | - | Variable (to 50 µL) | - |
| PCR Buffer | 10X | 5 µL | 1X |
| dNTP Mix | 10 mM (total) | 1 µL | 200 µM (each) |
| Forward Primer | 20 µM | 1 µL | 0.4 µM |
| Reverse Primer | 20 µM | 1 µL | 0.4 µM |
| DNA Template | Variable (e.g., 10 ng/µL) | 1 µL | e.g., 0.2 ng/µL |
| DNA Polymerase | 5 U/µL | 0.5 µL | 2.5 U |
| MgCl₂ | 25 mM | See Table 2 | Variable (1.5-5.0 mM) |
Table 2: MgCl₂ volumes for establishing a concentration gradient.
| Final [MgCl₂] (mM) | 1.5 | 2.0 | 2.5 | 3.0 | 3.5 | 4.0 | 4.5 | 5.0 |
|---|---|---|---|---|---|---|---|---|
| Volume of 25 mM MgCl₂ (µL) | 3.0 | 4.0 | 5.0 | 6.0 | 7.0 | 8.0 | 9.0 | 10.0 |
Protocol: MgCl₂ Optimization Experiment
- Prepare Master Mix: Calculate the required number of reactions (samples + controls + ~10% extra). In a sterile tube, combine all reagents from Table 1 except for the DNA template and MgCl₂ [8] [18]. Mix thoroughly by pipetting.
- Aliquot and Add MgCl₂: Distribute the Master Mix into individual PCR tubes. Add the corresponding volume of 25 mM MgCl₂ stock to each tube as per Table 2.
- Add Template: Add the DNA template to each sample tube. Add an equivalent volume of sterile water to the negative control tube.
- Thermal Cycling: Run the PCR using your standard cycling protocol. If possible, use a gradient thermal cycler to simultaneously test different annealing temperatures.
- Analysis: Analyze the PCR products using agarose gel electrophoresis. The optimal condition is the lowest MgCl₂ concentration that produces a strong, specific band of the expected size with no background smearing.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential reagents for PCR optimization with MgCl₂.
| Reagent / Material | Function / Rationale |
|---|---|
| MgCl₂ Stock Solution (25 mM) | Source of divalent Mg²⁺ ions, a essential cofactor for DNA polymerase activity. Its concentration directly influences priming efficiency, specificity, and amplicon yield. |
| Hot-Start DNA Polymerase | A modified enzyme inactive at room temperature, preventing nonspecific primer extension and primer-dimer formation during reaction setup, thereby enhancing specificity [11]. |
| Molecular Biology Grade Water | A nuclease-free, sterile water used to reconstitute primers and adjust reaction volume, preventing enzymatic degradation and introduction of contaminants. |
| dNTP Mix | The building blocks (dATP, dCTP, dGTP, dTTP) for DNA synthesis. Unbalanced dNTP concentrations can increase error rates; they also chelate Mg²⁺, affecting its free concentration [11]. |
| 10X PCR Buffer | Provides the optimal chemical environment (pH, ionic strength) for the DNA polymerase. Many buffers come with or without a pre-added MgCl₂ solution. |
Workflow and Relationship Diagrams
The diagram below illustrates the logical relationship between MgCl₂ concentration, its biochemical effects, and the final PCR outcome, providing a framework for troubleshooting.
Diagram: The cause-and-effect relationship between MgCl₂ concentration and PCR results.
A Step-by-Step Guide to Optimizing MgCl2 Concentration in Your PCR Protocol
FAQs on Magnesium Concentration in PCR
1. What is the fundamental role of MgCl₂ in a PCR reaction?
MgCl₂ is an essential cofactor for DNA polymerase enzymes. The magnesium ion (Mg²⁺) facilitates the catalytic activity of the enzyme, enabling the formation of phosphodiester bonds between nucleotides to create the new DNA strand. Additionally, Mg²⁺ neutralizes the negative charge on the phosphate backbone of DNA, which stabilizes the DNA duplex and facilitates proper primer binding by increasing the primer's melting temperature (Tm) [1].
2. What is the standard working range for MgCl₂ concentration?
For standard PCR reactions, the optimal concentration of MgCl₂ typically falls within a range of 1.5 mM to 5.0 mM [1] [19]. A comprehensive meta-analysis of optimization studies identified an optimal range of 1.5–3.0 mM for efficient PCR performance [4]. Most standard protocols often start with a concentration of around 2.0 mM [1].
3. How does MgCl₂ concentration specifically influence the formation of nonspecific PCR bands?
The MgCl₂ concentration is a critical determinant for amplification specificity.
- Too much MgCl₂ (e.g., >3-4 mM, depending on the reaction): Excess Mg²⁺ stabilizes DNA duplexes to the point where primers can bind to non-target, partially homologous sequences on the DNA template. This promiscuous binding leads to the amplification of unwanted DNA fragments, which appear as multiple bands or a smear on an agarose gel [11] [1] [19]. High Mg²⁺ concentrations can also reduce enzyme fidelity, increasing the chance of misincorporation [11] [20].
- Too little MgCl₂ (e.g., <1.5 mM): Insufficient Mg²⁺ results in poor DNA polymerase activity and unstable primer-template binding. This leads to weak or non-existent amplification of even the desired target product [1] [19].
4. How should I optimize MgCl₂ concentration to eliminate nonspecific bands in my experiment?
Begin by performing a MgCl₂ titration experiment. Set up a series of identical PCR reactions, varying only the MgCl₂ concentration. A recommended starting range is 0.5 mM to 5.0 mM in increments of 0.5 mM. Analyze the results using agarose gel electrophoresis to identify the concentration that yields a single, strong band of the expected size with the least background [11] [20]. This empirical approach is the most reliable way to determine the optimal condition for your specific primer-template system.
5. Do different DNA templates require different MgCl₂ concentrations?
Yes, template characteristics significantly influence the optimal MgCl₂ concentration. The meta-analysis revealed that template complexity is a key factor, with genomic DNA templates often requiring higher MgCl₂ concentrations than simpler templates like plasmids [4]. Furthermore, GC-rich templates, which form stable secondary structures, may require optimization of Mg²⁺, sometimes in combination with PCR enhancers like DMSO or betaine, to achieve efficient denaturation and amplification [11] [20] [6].
6. Besides MgCl₂, what other factors can cause nonspecific amplification?
Nonspecific amplification is often multifactorial. Other common causes include:
- Suboptimal Annealing Temperature: An annealing temperature that is too low is a primary cause of mispriming [11] [8].
- Excessive DNA Polymerase or Primers: High concentrations of enzyme or primers can promote off-target binding [11].
- Poor Primer Design: Primers with self-complementarity (leading to hairpins) or complementarity to each other (leading to primer-dimers) can cause nonspecific products [11] [8].
- High Cycle Number: An excessive number of PCR cycles can amplify low-level nonspecific products formed in earlier cycles [11] [21].
Table 1: Summary of Key Quantitative Findings on MgCl₂ in PCR
| Aspect | Quantitative Finding | Source |
|---|---|---|
| General Optimal Range | 1.5 - 5.0 mM | [1] [19] |
| Meta-Analysis Optimal Range | 1.5 - 3.0 mM | [4] |
| Common Starting Concentration | ~2.0 mM | [1] |
| Effect on DNA Melting Temperature (Tm) | Every 0.5 mM increase in MgCl₂ raises DNA Tm by ~1.2°C | [4] |
| Effect on Specificity | Concentrations >3.0 mM can increase nonspecific binding and reduce fidelity | [11] [4] [20] |
Experimental Protocol: MgCl₂ Titration for Optimization
This protocol provides a detailed methodology for empirically determining the optimal MgCl₂ concentration for your PCR assay.
1. Reagents and Materials
- Template DNA (e.g., genomic DNA)
- Forward and Reverse Primers
- 10X PCR Buffer (without MgCl₂)
- MgCl₂ stock solution (e.g., 25 mM)
- dNTP Mix
- DNA Polymerase
- Nuclease-free Water
- PCR Tubes or Plates
- Thermal Cycler
- Agarose Gel Electrophoresis Equipment
2. Procedure
Step 1: Prepare the Master Mix Create a master mix for n+1 reactions to minimize pipetting error. For a 50 µl reaction volume, calculate the total volumes needed for all components except MgCl₂ and template DNA.
Step 2: Aliquot Master Mix Dispense equal volumes of the master mix into a series of labeled PCR tubes.
Step 3: Titrate MgCl₂ Add MgCl₂ from a stock solution to each tube to achieve the desired final concentration range. A standard titration series is: 1.0 mM, 1.5 mM, 2.0 mM, 2.5 mM, 3.0 mM, 3.5 mM, 4.0 mM, and 5.0 mM.
Step 4: Add Template and Initiate PCR Add a consistent amount of template DNA to each tube. Gently mix and briefly centrifuge to collect the contents. Place the tubes in a thermal cycler and run the standard PCR program optimized for your primers and expected amplicon size.
Step 5: Analyze Results Separate the PCR products by agarose gel electrophoresis. Visualize the bands under UV light. The optimal MgCl₂ concentration is the one that produces the most intense, specific band of the correct size with the least background smear or extra bands.
Workflow Diagram for PCR Optimization
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for PCR Optimization with MgCl₂
| Reagent | Function | Considerations for Optimization |
|---|---|---|
| MgCl₂ (Magnesium Chloride) | Essential cofactor for DNA polymerase; stabilizes primer-template binding and influences DNA duplex stability [1]. | The single most critical variable to titrate. Directly controls reaction specificity and efficiency [11] [4]. |
| Hot-Start DNA Polymerase | Enzyme engineered to be inactive at room temperature, preventing nonspecific amplification and primer-dimer formation during reaction setup [11]. | Using a hot-start enzyme is a best practice that provides a cleaner baseline from which to optimize Mg²⁺. |
| PCR Buffer (without MgCl₂) | Provides the ionic environment (e.g., Tris-HCl for pH, KCl) and salts necessary for robust enzyme activity [8] [20]. | Using a Mg-free buffer is essential for a precise titration experiment. |
| PCR Enhancers (DMSO, Betaine) | Additives that help denature complex DNA templates, especially those with high GC content, by reducing secondary structure formation [11] [20] [6]. | Often used in conjunction with Mg²⁺ optimization for challenging templates. May require re-optimization of Mg²⁺ concentration. |
| dNTP Mix | The building blocks (dATP, dCTP, dGTP, dTTP) for the new DNA strands [8]. | dNTPs chelate Mg²⁺. Ensure dNTP concentrations are consistent and balanced, as changes will affect the amount of free Mg²⁺ available [11]. |
FAQ: Why am I seeing multiple bands or a smeared background in my PCR gel?
A: Nonspecific amplification, resulting in multiple bands or a smear, is a common issue often caused by suboptimal reaction stringency. Key factors include:
- Annealing Temperature: If the temperature is too low, primers can bind to incorrect, partially complementary sites on the DNA template [22].
- MgCl₂ Concentration: Excess Mg²⁺ can reduce reaction fidelity and promote the amplification of nonspecific products [11] [22].
- Template Quality and Quantity: Degraded DNA or too much template DNA can lead to smearing and high background [11] [23].
- Primer Design and Concentration: Poorly designed primers or excessively high primer concentrations can cause primer-dimer formation and nonspecific binding [11] [24].
A systematic approach using Gradient PCR and MgCl₂ Titration is the most effective way to identify the precise conditions that suppress these artifacts.
FAQ: What is a systematic workflow for optimizing PCR conditions?
A robust optimization protocol involves testing one variable at a time while keeping others constant. The following workflow outlines this systematic approach.
Experimental Protocol: Gradient PCR for Annealing Temperature Optimization
This experiment determines the optimal annealing temperature (T_a) for your specific primer-template combination in a single run [25] [22].
1. Prepare Master Mix:
Create a master mix for n+1 reactions (where n is the number of gradient wells you will use). The table below outlines a standard 50 µL reaction volume.
Table: PCR Master Mix Components
| Reagent | Final Concentration | Volume per 50 µL Reaction |
|---|---|---|
| 10X PCR Buffer | 1X | 5 µL |
| dNTP Mix | 200 µM each | 1 µL (from 10 mM stock) |
| Forward Primer | 0.5 µM | 1.25 µL (from 20 µM stock) |
| Reverse Primer | 0.5 µM | 1.25 µL (from 20 µM stock) |
| MgCl₂ (25 mM) | 1.5 mM (starting point) | 3 µL |
| DNA Template | ~100 ng (genomic) | Variable |
| DNA Polymerase | 1.25 U | 0.25 µL (from 5 U/µL stock) |
| Nuclease-Free Water | - | To 50 µL |
2. Aliquot and Run Gradient PCR:
- Aliquot the master mix into
nPCR tubes or a 96-well plate. - Program your thermal cycler with a gradient annealing step. Set the gradient to span a range, typically 5°C below to 5°C above the calculated theoretical
T_m(melting temperature) of your primers [25] [26]. - Run the PCR program.
Table: Example Thermal Cycler Program
| Step | Temperature | Time | Cycles |
|---|---|---|---|
| Initial Denaturation | 94-98°C | 1-5 minutes | 1 |
| Denaturation | 94-98°C | 10-30 seconds | |
| Annealing (Gradient) | Variable (e.g., 55-65°C) | 30 seconds | 30-35 |
| Extension | 68-72°C | 1 minute/kb | |
| Final Extension | 68-72°C | 5-10 minutes | 1 |
| Hold | 4-10°C | ∞ | 1 |
3. Analyze Results:
- Analyze the PCR products using agarose gel electrophoresis.
- Identify the well/temperature that produces a single, sharp band of the expected size with the least background smearing. This is your optimal annealing temperature [25].
Experimental Protocol: MgCl₂ Titration
Once the optimal T_a is found, perform an MgCl₂ titration to further enhance specificity and yield [11] [10].
1. Prepare Titration Master Mix:
Prepare a master mix identical to the one above but omit MgCl₂. Aliquot this Mg-free master mix into a series of n tubes.
2. Add MgCl₂: Add MgCl₂ (from a stock solution, e.g., 25 mM) to each tube to create a concentration series. A typical range is 0.5 mM to 5.0 mM [26].
Table: Example MgCl₂ Titration Series
| Tube | MgCl₂ (25 mM Stock) | Final [MgCl₂] in 50 µL |
|---|---|---|
| 1 | 1.0 µL | 0.5 mM |
| 2 | 2.0 µL | 1.0 mM |
| 3 | 3.0 µL | 1.5 mM |
| 4 | 4.0 µL | 2.0 mM |
| 5 | 6.0 µL | 3.0 mM |
| 6 | 8.0 µL | 4.0 mM |
| 7 | 10.0 µL | 5.0 mM |
3. Run PCR and Analyze:
- Run the PCR using the optimized annealing temperature determined from the gradient experiment.
- Analyze the results by gel electrophoresis. The ideal MgCl₂ concentration will yield the brightest specific band with the cleanest background.
The relationship between MgCl₂ concentration and PCR results is summarized below.
The Scientist's Toolkit: Key Research Reagent Solutions
The following table details essential reagents for PCR optimization experiments, highlighting their role in reducing nonspecific amplification.
Table: Essential Reagents for PCR Optimization
| Reagent | Function in PCR | Optimization Role & Impact on Specificity |
|---|---|---|
| Hot-Start DNA Polymerase | Enzyme that synthesizes new DNA strands. "Hot-start" versions are inactive until a high-temperature activation step. | Critical. Prevents primer-dimer formation and nonspecific priming during reaction setup by inhibiting enzyme activity at low temperatures [11] [10] [24]. |
| Magnesium Chloride (MgCl₂) | Essential cofactor for DNA polymerase activity. Stabilizes primer-template binding [27]. | Primary optimization target. Concentration directly influences specificity; too little reduces yield, too much promotes nonspecific binding and reduces fidelity [11] [10] [26]. |
| Ultrapure dNTPs | The building blocks (dATP, dCTP, dGTP, dTTP) for DNA synthesis. | Required at balanced equimolar concentrations. Unbalanced dNTPs can increase error rates and favor misincorporation [11] [27]. |
| PCR Additives (e.g., DMSO, BSA, Betaine) | Modifies DNA melting behavior and polymerase stability. | Secondary optimization. DMSO can help denature GC-rich templates. BSA can bind inhibitors. Use the lowest effective concentration to improve specificity of difficult amplifications [11] [27]. |
| Gradient Thermal Cycler | Instrument that allows different wells to run at different temperatures simultaneously during the annealing step. | Enables efficient optimization. Allows for the empirical determination of the optimal annealing temperature in a single experiment, saving time and reagents [25]. |
FAQs: The Magnesium Balance in PCR
FAQ 1: Why is free Mg2+ concentration critical for PCR specificity and efficiency? Magnesium ions (Mg2+) are an essential cofactor for DNA polymerase activity. The enzyme requires free Mg2+ to catalyze the formation of phosphodiester bonds between nucleotides. An incorrect concentration can lead to two primary issues: insufficient Mg2+ results in low PCR yield or failed amplification, while excess Mg2+ decreases specificity and promotes the formation of nonspecific bands and primer-dimers by stabilizing nonspecific primer-template interactions [28].
FAQ 2: How do dNTPs and EDTA chelate free Mg2+ ions in a PCR? Both dNTPs and EDTA act as Mg2+ chelators by binding the ions and rendering them unavailable for the DNA polymerase.
- dNTPs: The phosphate groups of dNTPs bind Mg2+ to form biologically active complexes (e.g., Mg-dNTP). The polymerase uses these complexes as substrates. The concentration of dNTPs in a reaction is directly proportional to the amount of Mg2+ they chelate.
- EDTA: Ethylenediaminetetraacetic acid (EDTA) is a potent chelating agent often present in DNA elution buffers or used in sample preparation. It strongly binds to divalent cations like Mg2+, and even low concentrations can completely sequester Mg2+, leading to PCR failure [29].
FAQ 3: How can I calculate the optimal concentration of MgCl2 for my reaction? A common starting point is to use a concentration of Mg2+ that is in excess of the total dNTP concentration. A standard rule of thumb is provided in the table below. However, because other reaction components can also affect Mg2+ availability, empirical optimization is necessary.
Table 1: Guideline for Mg2+ to dNTP Molar Ratio
| Parameter | Typical Concentration in PCR | Function / Interaction with Mg2+ |
|---|---|---|
| Free Mg2+ | 0.5 - 5.0 mM (optimization required) | Essential DNA polymerase cofactor. |
| dNTPs (each) | 0.2 - 0.5 mM | Chelates Mg2+ (as Mg-dNTP substrate). |
| EDTA | Should be minimized (< 0.1 mM) | Potently chelates and inactivates Mg2+. |
The following formula can be used as an initial guide:
[Mg2+]_free ≈ [Mg2+]_total - [α * dNTP_total]
Where α is a coefficient representing the binding ratio (typically between 0.8 and 1). This highlights that a significant portion of the total Mg2+ is bound to dNTPs and is not "free." Therefore, the total MgCl2 added must be high enough to satisfy the chelation by dNTPs and still provide a sufficient concentration of free Mg2+ for the polymerase.
FAQ 4: What are the signs of Mg2+-related issues in my PCR results?
- No Amplification: Could indicate complete Mg2+ chelation, often due to EDTA contamination or excessively low MgCl2.
- Nonspecific Bands/Smearing: Often a sign of excessively high MgCl2 concentration, which reduces reaction stringency.
- Low Yield: Can result from insufficient free Mg2+ for optimal polymerase activity.
Troubleshooting Guide: Resolving Mg2+ Imbalance
Table 2: Troubleshooting Mg2+, dNTP, and EDTA Issues
| Problem Observed | Potential Cause | Recommended Solution |
|---|---|---|
| No PCR product | EDTA contamination from DNA template or buffers. | Ensure EDTA concentration in the final reaction is < 0.1 mM. Use a DNA template purified with EDTA-free buffers or dilute the template. |
| Excessively low MgCl2 concentration. | Perform a MgCl2 titration, increasing the concentration in 0.5 mM increments from 0.5 mM to 5.0 mM. | |
| Multiple nonspecific bands or smearing | Excess free Mg2+ leading to low-fidelity amplification. | Perform a MgCl2 titration, decreasing the concentration in 0.5 mM increments. |
| High dNTP concentration inconsistently chelating Mg2+. | Use a consistent, standardized concentration of dNTPs (e.g., 0.2 mM each). | |
| Inconsistent results between replicates | Variable carryover of EDTA from sample prep. | Standardize DNA isolation and purification methods. Use a chelator-resistant polymerase (see Toolkit). |
| Inaccurate pipetting of concentrated MgCl2 stock. | Create a ready-to-use PCR master mix to minimize pipetting error. |
Experimental Protocol: Optimizing MgCl2 Concentration
This protocol provides a methodology for empirically determining the optimal MgCl2 concentration to reduce nonspecific bands, framed within a thesis research context.
Aim: To determine the MgCl2 concentration that maximizes specific product yield and minimizes nonspecific amplification for a given primer-template system.
Background: The theoretical starting MgCl2 concentration can be calculated based on dNTP concentration, but empirical validation is crucial due to the influence of other reaction components, such as primers and template, which can also weakly bind Mg2+ [28]. Furthermore, amplifying difficult templates like GC-rich regions often requires deviation from standard conditions [28].
Materials:
- See "The Scientist's Toolkit" below.
- Template DNA (e.g., genomic DNA).
- Target-specific primers.
- Thermostable DNA polymerase with corresponding reaction buffer (without MgCl2).
- PCR tubes/plates and thermal cycler.
Method:
- Prepare Master Mix: Create a master mix for
n+1reactions (wherenis the number of MgCl2 conditions) containing nuclease-free water, reaction buffer (without MgCl2), primers, dNTP mix, DNA polymerase, and template DNA. Mix thoroughly by gentle vortexing and brief centrifugation. - Aliquot: Dispense equal volumes of the master mix into
nPCR tubes. - Spike MgCl2: Add MgCl2 stock solution to each tube to create a titration series. A typical range is 1.0 mM to 4.0 mM in 0.5 mM increments.
- Example: Tube 1: 1.0 mM MgCl2; Tube 2: 1.5 mM; Tube 3: 2.0 mM; ... Tube 7: 4.0 mM.
- Run PCR: Place the tubes in a thermal cycler and start the optimized amplification program.
- Analyze Results: Separate the PCR products using agarose gel electrophoresis (e.g., 2% agarose gel) [28] [29].
- Identify the optimal condition: The MgCl2 concentration that produces a single, intense band of the expected size with minimal or no nonspecific bands or primer-dimer is considered optimal.
Diagram 1: MgCl2 optimization workflow.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Managing Mg2+ in PCR
| Reagent / Material | Function / Role in Mg2+ Context | Considerations for Use |
|---|---|---|
| MgCl2 Stock Solution | Source of divalent Mg2+ ions. | Concentration must be accurately known. Titration is mandatory for assay optimization. |
| dNTP Mix | Provides nucleotides (dATP, dCTP, dGTP, dTTP) for DNA synthesis. | Chelates Mg2+. Keep concentration constant during Mg2+ titration to avoid confounding variables. |
| EDTA-free Buffers | For DNA template purification and resuspension. | Prevents introduction of a potent Mg2+ chelator into the PCR [29]. |
| Chelator-Resistant Polymerase (e.g., KOD) | DNA polymerase engineered for tolerance to common inhibitors. | KOD polymerase has been shown to be more resistant to metal ion inhibition compared to Taq polymerase [29]. |
| PCR Additives (e.g., DMSO) | Assist in amplifying difficult templates (e.g., GC-rich). | Additives like DMSO can change template accessibility and may slightly alter the optimal Mg2+ concentration [28]. |
Diagram 2: Mg2+ interactions and PCR outcomes.
FAQs and Troubleshooting Guides
▸ What is the core principle behind combining Touchdown PCR and MgCl2 optimization?
This combination uses a multi-pronged strategy to maximize PCR specificity. Touchdown PCR begins with an annealing temperature several degrees above the primers' calculated melting temperature (Tm). This high initial stringency ensures that only the most perfectly matched primer-template hybrids form, effectively suppressing non-specific amplification from the very first cycles. The annealing temperature is then gradually decreased—typically by 1°C per cycle—until the optimal Tm is reached. This allows the desired specific product, which has been preferentially amplified in the early cycles, to outcompete any potential non-specific products in later, more permissive cycles.
MgCl2 optimization works synergistically with this approach. Magnesium ions (Mg²⁺) are an essential cofactor for DNA polymerase, and their concentration directly affects enzyme fidelity and primer annealing. Excessive Mg²⁺ can reduce enzyme fidelity and promote non-specific primer binding, while insufficient Mg²⁺ can lead to poor polymerase activity and low yield. By carefully calibrating the Mg²⁺ concentration, you create an optimal chemical environment that supports the high-fidelity amplification promoted by the touchdown thermal profile, leading to a dramatic reduction in nonspecific bands [30] [31] [32].
▸ My gel still shows multiple bands after using a standard Touchdown PCR protocol. What should I do next?
Persistent multiple bands indicate that the reaction conditions require further optimization. You should systematically address the key variables. Follow the troubleshooting workflow below to diagnose and resolve the issue:
First, re-evaluate your primer design using multiple tools like NCBI Primer-BLAST to check for off-target binding sites and ensure there are no self-complementary regions or hairpins [7]. If primers are correct, begin a MgCl₂ titration. Since Mg²⁺ concentration is critical, prepare a series of reactions testing a range from 1.0 mM to 4.0 mM in 0.5 mM increments. This will help you identify the concentration that provides the best specificity for your specific primer-template combination [31] [8] [33].
If bands persist, incorporate PCR enhancers. Additives like DMSO (1-10%), betaine (0.5 M to 2.5 M), or BSA (10-100 μg/mL) can help disrupt secondary structures and stabilize the polymerase, particularly with difficult templates [31] [8] [33]. Finally, confirm you are using a hot-start polymerase to prevent primer-dimer formation and non-specific amplification during reaction setup [34] [33].
▸ How do I determine the correct starting and ending annealing temperatures for Touchdown PCR?
The temperature parameters for Touchdown PCR are based on the melting temperature (Tm) of your primer set.
- Calculate the Tm of your primers: Use the formula: Tm = 2(A + T) + 4(G + C), where A, T, G, and C represent the number of each base in the primer. Calculate this for both forward and reverse primers [30].
- Identify the lowest Tm: Use the lower Tm value from your two primers for a conservative approach.
- Set the starting (highest) annealing temperature: Begin 8-10°C above the lowest primer Tm. For example, if your lowest Tm is 57°C, start your first cycles at 67°C [35].
- Set the final (lowest) annealing temperature: This is typically 3-5°C below the lowest primer Tm. In our example, this would be 52-54°C [30] [35].
The following protocol provides a detailed example of how to implement this:
Table: Example Touchdown PCR Protocol Based on a Primer Tm of 57°C
| Step | Temperature (°C) | Time | Stage and Number of Cycles |
|---|---|---|---|
| 1. Initial Denaturation | 95 | 3:00 | |
| 2. Denature | 95 | 0:30 | Stage 1: 10 cycles |
| 3. Anneal | 67 (Tm +10) | 0:45 | |
| 4. Extension | 72 | 0:45 | |
| 5. Denature | 95 | 0:30 | Stage 2: 15-20 cycles |
| 6. Anneal | 57 (Calculated Tm) | 0:45 | |
| 7. Extension | 72 | 0:45 | |
| 8. Final Extension | 72 | 5:00 |
Note: In Stage 1, the annealing temperature decreases by 1°C per cycle from 67°C down to 57°C over the 10 cycles. Stage 2 then continues for the remaining cycles at the final annealing temperature of 57°C [35].
▸ I get no amplification product at all. How can I determine if MgCl₂ is the problem?
A complete absence of product suggests a failure in the reaction's core components. To diagnose if MgCl₂ is the issue, follow a systematic approach. First, run a positive control with a primer and template combination that is known to work under your standard PCR conditions. If the positive control fails, the problem likely lies with your master mix or thermocycler, not specifically with MgCl₂.
If the positive control works, then perform a MgCl₂ titration series. Prepare reactions testing a range of MgCl₂ concentrations, for example: 0.5 mM, 1.0 mM, 1.5 mM, 2.0 mM, 2.5 mM, 3.0 mM, 3.5 mM, and 4.0 mM. The presence of a band in one or more of these tubes will immediately reveal both whether Mg²⁺ is the limiting factor and what the optimal concentration is for your assay [31] [8].
Also, consider other common causes of failed amplification:
- Too little template: Use an appropriate amount (e.g., 10-100 ng of genomic DNA) [30] [32].
- Incorrect annealing temperature: The final touchdown temperature might be too high. Verify your Tm calculations and consider lowering the final annealing temperature by a few degrees [33].
- Template quality: The DNA could be degraded or contain inhibitors. Re-purify your template or try a dilution series to dilute out potential inhibitors [31] [33].
▸ Are there specific considerations for combining these techniques with GC-rich templates?
Yes, GC-rich templates (>65% GC content) present unique challenges due to their stable secondary structures and high melting temperatures, but the combination of Touchdown PCR and MgCl₂ optimization is particularly well-suited to overcome them.
Modifications to the Standard Protocol:
- Higher Denaturation Temperature: Use 98°C instead of 94-95°C for the denaturation step to ensure complete separation of the tightly bound DNA strands [34] [32].
- PCR Additives: The use of additives is highly recommended. DMSO (at 2.5-5%) or betaine (1.5 M) are very effective at reducing secondary structure formation in GC-rich regions, which greatly improves amplification efficiency and specificity [34] [32] [36].
- Polymerase Choice: Consider using a polymerase specifically engineered for high GC content, as they are often more processive and can better "power through" difficult secondary structures [30] [34] [32].
Table: Optimized Reaction Components for GC-Rich Templates
| Component | Standard Recommendation | GC-Rich Optimization |
|---|---|---|
| Initial Denaturation | 94-95°C for 1-3 min | 98°C for 2-5 min |
| Denaturation | 94-95°C for 15-30 sec | 98°C for 10-15 sec |
| MgCl₂ | Titrate 1.5-2.0 mM | Titrate 2.0-3.0 mM (may require higher concentration) |
| Additives | Often not required | DMSO (2.5-5%), Betaine (0.5-1.5 M) |
| DNA Polymerase | Standard Taq | Specialized polymerase for GC-rich templates |
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Reagents for Touchdown PCR and MgCl₂ Optimization
| Item | Function | Brief Explanation & Application Note |
|---|---|---|
| Hot-Start DNA Polymerase | Reduces nonspecific amplification during reaction setup. | An enzyme chemically modified or bound by an antibody to be inactive at room temperature. Activated during the initial denaturation step. Critical for maintaining the specificity gains of Touchdown PCR [34]. |
| MgCl₂ Solution (25-50 mM) | Essential cofactor for DNA polymerase activity. | The optimal concentration is template- and primer-specific. Must be titrated to find the concentration that balances yield and specificity. Supplied separately from the buffer for optimization [31] [32]. |
| PCR Additives (DMSO, Betaine, BSA) | Disrupt DNA secondary structures, stabilize enzymes, or bind inhibitors. | DMSO: Helps denature GC-rich templates. Use at 1-10%. Betaine: Equalizes the melting temperature of AT- and GC-rich regions. Use at 0.5-2.5 M. BSA: Binds to inhibitors in the template prep. Use at 10-100 μg/mL [31] [8] [33]. |
| dNTP Mix | Building blocks for new DNA synthesis. | Typically used at 50-200 μM of each dNTP. Too high a concentration can decrease specificity and chelate Mg²⁺, thereby reducing the free [Mg²⁺] available for the polymerase [30] [8]. |
| Nuclease-Free Water | Solvent for the reaction. | Used to bring the reaction to its final volume. Must be nuclease-free to prevent degradation of primers, template, and reagents [8]. |
| Gradient Thermal Cycler | Allows empirical determination of optimal annealing temperature. | Enables you to test a range of annealing temperatures (e.g., for the final touchdown step or for standard PCR optimization) in a single run, saving time and reagents [30]. |
The Critical Role of MgCl2 in PCR Specificity
Why is MgCl2 concentration so pivotal for preventing nonspecific amplification?
Magnesium chloride (MgCl₂) is an essential cofactor in the Polymerase Chain Reaction (PCR) because it directly influences the activity of DNA polymerase and the stability of the newly synthesized DNA duplex. Its concentration is a key determinant in the success of an experiment, especially when aiming to reduce nonspecific bands [13]. Mg²⁺ ions facilitate the formation of a soluble complex with dNTPs, which is a prerequisite for their incorporation into the growing DNA strand by the polymerase [37]. Furthermore, they act as a necessary co-factor for the enzymatic activity of Taq polymerase and stabilize the primer-template interaction by increasing its melting temperature (Tm) [37].
An imbalance in MgCl₂ concentration is a common source of PCR artifacts. Too little MgCl₂ results in reduced polymerase activity, leading to weak or non-existent amplification of the desired target [38]. Conversely, too much MgCl₂ can promote non-specific primer binding, which manifests on an agarose gel as multiple bands, smears, or a ladder of DNA products [8] [38]. This happens because excess Mg²⁺ stabilizes even weak, incorrect primer-template interactions, allowing amplification from non-target sites. Therefore, fine-tuning the MgCl₂ concentration is a fundamental strategy for enhancing specificity and achieving a single, strong band corresponding to the target amplicon.
Quantitative Guidelines for MgCl2 Optimization
What are the evidence-based optimal ranges for MgCl2?
Extensive research, including a recent systematic meta-analysis of 61 studies, has provided quantitative insights into the effects of MgCl₂ [4] [15]. The optimal concentration is not a single value but a range that must be tailored to the specific reaction components and template characteristics.
The table below summarizes the general and template-specific guidelines for MgCl₂ concentration:
| Template Type | Recommended Starting Point | Optimal Range (for many templates) | Key Considerations |
|---|---|---|---|
| Standard Templates | 1.5 mM [8] | 1.5 - 3.0 mM [4] [15] | A standard starting point included in many commercial buffers. |
| GC-Rich Templates | 2.0 mM [13] | May require up to 4.0 mM [38] | Higher Mg²⁺ helps denature stable secondary structures. Test in 0.5 mM increments [38]. |
| Complex Genomic DNA | 2.0 mM | Higher concentrations often required [4] [15] | Increased complexity and size of the genome can demand more Mg²⁺ for efficient polymerization. |
A key quantitative finding is the logarithmic relationship between MgCl₂ concentration and DNA melting temperature. Within the 1.5 to 3.0 mM range, every 0.5 mM increase in MgCl₂ raises the DNA melting temperature by approximately 1.2°C [4] [15]. This directly impacts the primer annealing efficiency and must be considered when setting the annealing temperature.
Research Reagent Solutions
The following table details key reagents used in the optimization of MgCl₂ for challenging PCRs:
| Reagent / Solution | Function in PCR Optimization |
|---|---|
| MgCl₂ Stock Solution | Provides the Mg²⁺ ions essential for polymerase activity, dNTP complex formation, and primer-template stability [38] [37]. |
| dNTP Mix | The building blocks for DNA synthesis. Concentration is critical as dNTPs can chelate Mg²⁺ ions, effectively reducing the free Mg²⁺ available for the polymerase [37]. |
| PCR Buffer (with & without Mg²⁺) | Provides the optimal ionic environment and pH for the reaction. Using a buffer without pre-added MgCl₂ allows for flexible and precise optimization of Mg²⁺ concentration. |
| GC Enhancers (e.g., DMSO, Betaine) | Additives that reduce the formation of secondary structures in GC-rich templates, facilitating polymerase progression and improving yield and specificity [38]. |
| Thermostable DNA Polymerase | The enzyme that catalyzes DNA synthesis. Specialized polymerases (e.g., Q5, OneTaq) are often more effective at amplifying difficult templates like GC-rich sequences [38]. |
Experimental Protocol: A Step-by-Step Optimization Guide
How do I systematically optimize MgCl2 concentration for my specific template?
This protocol provides a detailed methodology for empirically determining the ideal MgCl₂ concentration to minimize nonspecific bands.
Materials and Reagents
- Template DNA (e.g., genomic DNA, plasmid)
- Forward and Reverse Primers
- 10X PCR Buffer (without MgCl₂)
- MgCl₂ stock solution (e.g., 25 mM or 50 mM)
- dNTP mix
- DNA Polymerase
- Nuclease-free water
- PCR tubes and thermal cycler
Procedure
Prepare a Master Mix: Create a master mix containing all common reagents for the number of reactions you are testing, plus a 10% excess to account for pipetting error. For a single 50 µl reaction, the core components are:
- 5.0 µl of 10X PCR Buffer (without MgCl₂)
- 1.0 µl of 10 mM dNTP mix (final 200 µM each)
- 1.0 µl of Forward Primer (20 µM stock)
- 1.0 µl of Reverse Primer (20 µM stock)
- 0.5-2.5 Units of DNA Polymerase (see manufacturer's recommendation)
- 1-1000 ng of Template DNA
- X µl of MgCl₂ stock solution (variable)
- Nuclease-free water to a final volume of 50 µl [8]
Set Up the MgCl₂ Gradient: Aliquot the master mix into individual PCR tubes. Then, add MgCl₂ stock solution to each tube to achieve a final concentration gradient. A typical optimization might include the following final concentrations:
Run the PCR: Place the tubes in a thermal cycler and start the PCR program. If available, use a gradient function to simultaneously test different annealing temperatures, as the optimal MgCl₂ concentration and annealing temperature are interdependent [37].
Analyze the Results: Separate the PCR products by agarose gel electrophoresis. Identify the MgCl₂ concentration that produces a single, intense band of the expected size with minimal to no background smearing or non-specific bands.
The following workflow diagram outlines the logical steps for this optimization process:
Advanced Strategies and FAQs
How do I handle extremely challenging templates like GC-rich sequences?
For GC-rich templates (≥60% GC content), standard optimization may be insufficient. These sequences form highly stable secondary structures and require a multi-pronged approach [38].
- Polymerase Choice: Use polymerases specifically engineered for high GC content, such as Q5 or OneTaq, which often come with specialized GC buffers and enhancers [38].
- Combination with Additives: Incorporate PCR enhancers like DMSO, betaine, or glycerol. These compounds help denature secondary structures and increase primer stringency, working synergistically with optimized MgCl₂ [38].
- "Touchdown" PCR: This technique can be beneficial. It starts with a high annealing temperature to ensure maximum specificity in the first cycles and gradually lowers it to improve efficiency.
Frequently Asked Questions (FAQs)
Q1: My PCR worked with a published protocol. Should I still optimize MgCl₂? Even if a protocol works, optimization is recommended if you are transferring it to a new lab environment, using different reagent batches, or if the results show faint non-specific bands. Optimization ensures robustness and maximum specificity for your specific setup [37].
Q2: How does dNTP concentration relate to MgCl₂ optimization? dNTPs chelate Mg²⁺ ions. Therefore, the concentration of dNTPs directly affects the amount of free Mg²⁺ available for the polymerase. If you change the dNTP concentration, you must re-optimize the MgCl₂ concentration. A higher dNTP concentration generally requires a higher MgCl₂ concentration [37].
Q3: What is the most efficient way to optimize both MgCl₂ and annealing temperature? The most efficient method is to use a thermal cycler with a gradient function. This allows you to set up a single experiment where MgCl₂ concentration varies across one axis (e.g., different rows) and annealing temperature varies across the other (e.g., different columns) [37].
Troubleshooting Nonspecific Bands: A Systematic Approach to MgCl2 Adjustment
FAQ
What is the role of MgCl2 in a PCR reaction?
Magnesium chloride (MgCl2) is an essential cofactor for thermostable DNA polymerases. It serves two critical functions:
- Polymerase Cofactor: The Mg2+ ion is required for the catalytic activity of the DNA polymerase. It binds to a dNTP at its alpha phosphate group, facilitating the removal of beta and gamma phosphates and enabling the formation of a phosphodiester bond with the 3' OH group of the adjacent nucleotide [1].
- Nucleic Acid Stabilizer: Mg2+ ions bind to the negatively charged phosphate backbone of DNA. This reduces electrostatic repulsion between the primer and the template DNA strand, facilitating stable annealing and influencing the primer's melting temperature (Tm) [1] [3].
How can I tell if my MgCl2 concentration is too high?
Excessive MgCl2 concentration is a common cause of nonspecific amplification in PCR. The primary symptom observed during result analysis is:
- Multiple Bands or a DNA Smear on an agarose gel, instead of a single, sharp band of the expected size [13] [1]. This occurs because high Mg2+ concentrations reduce reaction stringency, allowing primers to anneal to non-target sites on the DNA template with partial complementarity [39] [13].
What is the typical optimal range for MgCl2 concentration?
The optimal MgCl2 concentration must be determined empirically for each primer-template system, but general guidelines are well-established:
| MgCl2 Concentration | Effect on PCR |
|---|---|
| Too Low (< 1.5 mM) | Weak or no amplification due to insufficient DNA polymerase activity and poor primer annealing [1]. |
| Optimal Range (1.5 - 3.0 mM) | Balanced specific activity and primer annealing, leading to specific amplification of the target sequence [4]. The most common starting concentration is 2.0 mM [1]. |
| Too High (> 3.0 mM) | Increase in nonspecific products and primer-dimer formation due to reduced primer annealing stringency; can also reduce enzyme fidelity [39] [4] [1]. |
For standard PCR, a concentration between 1.5 mM and 3.0 mM is often effective [4]. However, some specific primers or challenging templates (like GC-rich sequences) may require optimization up to 4.5 mM or slightly higher [40] [41].
Besides MgCl2, what other factors can cause multiple bands?
While MgCl2 is a key suspect, other reaction components and conditions can also lead to nonspecific amplification:
- Low Annealing Temperature: Reduces the stringency of primer binding [41].
- Poor Primer Design: Primers with self-complementarity, high GC 3' ends, or those that form primer-dimers can cause multiple bands [8] [3].
- Excessive Primer or Enzyme Concentration: Can promote mispriming and off-target synthesis [13] [3].
Troubleshooting Guide: A Step-by-Step Optimization Protocol
If you observe multiple bands, follow this systematic approach to determine if MgCl2 is the culprit and to identify the optimal concentration.
Step 1: Perform a MgCl2 Titration Experiment
The most direct method to optimize MgCl2 is to test a range of concentrations in a single experiment.
Materials Needed:
- Research Reagent Solutions
- Template DNA: High-quality genomic, cDNA, or plasmid DNA.
- Primers: Well-designed, highly specific oligonucleotides.
- MgCl2 Stock Solution: Typically 25 mM, provided with the DNA polymerase.
- 10X PCR Buffer (Mg-free): To control the initial Mg2+ concentration.
- dNTP Mix: Equimolar mixture of all four dNTPs.
- DNA Polymerase: Thermostable enzyme (e.g., Taq polymerase).
- Sterile Water: Nuclease-free to bring the reaction to final volume.
Detailed Methodology:
- Prepare a Master Mix containing all the common reagents for your number of reactions plus one extra to account for pipetting error. For a 50 µL reaction, the core components are:
- 5.0 µL of 10X PCR Buffer (Mg-free)
- 1.0 µL of 10 mM dNTP mix (final 200 µM of each dNTP)
- 1.0 µL of each primer (20 µM stock, final 0.4 µM each)
- 1.0 µL of DNA template (10-100 ng)
- 0.5 µL of DNA Polymerase (e.g., 2.5 U/µL)
- X µL of Sterile Water
- Y µL of 25 mM MgCl2 Stock (variable) [8]
- Aliquot the Master Mix into 8 individual PCR tubes.
- Add a different volume of the 25 mM MgCl2 stock to each tube to create a final concentration gradient. A recommended range is 1.0 mM to 4.0 mM in 0.5 mM increments [41]. Use the table below to calculate volumes for a 50 µL reaction.
Step 2: Calculate and Set Up Reactions
Use this table as a guide for setting up your titration experiment.
| Tube | Desired [MgCl2] (mM) | Volume of 25 mM MgCl2 Stock (µL) | Final Volume with Water (µL) |
|---|---|---|---|
| 1 | 1.0 | 2.0 | 50 |
| 2 | 1.5 | 3.0 | 50 |
| 3 | 2.0 | 4.0 | 50 |
| 4 | 2.5 | 5.0 | 50 |
| 5 | 3.0 | 6.0 | 50 |
| 6 | 3.5 | 7.0 | 50 |
| 7 | 4.0 | 8.0 | 50 |
| 8* | Varies | 0.0 | 50 |
Note: Tube 8 is a negative control without MgCl2 to confirm the requirement of Mg2+ for amplification.
Step 3: Run PCR and Analyze Results
- Place the tubes in a thermal cycler and run your standard PCR protocol.
- After cycling, analyze the PCR products using agarose gel electrophoresis.
- Identify the MgCl2 concentration that produces a single, strong band of the expected size with the least or no nonspecific bands. The relationship between MgCl2 and the results can be visualized as follows:
Advanced Considerations for Specific Cases
- GC-Rich Templates: DNA with high GC content (>60%) forms stable secondary structures that are difficult to denature. For these challenging templates, you may need to use a specialized polymerase and increase the MgCl2 concentration beyond the standard range, potentially up to 4.0 mM or higher, often in combination with additives like DMSO or betaine [41].
- Interacting Components: The concentration of free Mg2+ is critical, and it can be bound by other reagents, notably dNTPs. Ensure a balanced ratio, as excess dNTPs can chelate Mg2+, effectively reducing its availability for the polymerase [3]. A systematic meta-analysis has quantified that every 0.5 mM increase in MgCl2 within the 1.5-3.0 mM range is associated with an approximate 1.2 °C increase in DNA melting temperature, which directly affects primer annealing stringency [4].
FAQ: Magnesium Chloride and PCR Optimization
1. Why does high MgCl2 concentration cause primer-dimers and nonspecific bands? Magnesium chloride (MgCl2) is an essential cofactor for DNA polymerase activity. However, when the concentration is too high, it reduces the stringency of the PCR reaction. Excess Mg2+ stabilizes the binding between DNA strands, even when the match is not perfect. This allows primers to anneal to non-target sequences or to each other with greater ease, leading to the amplification of nonspecific products and the formation of primer-dimers [42] [11] [13]. Primer-dimers are short, artifactual double-stranded DNA fragments that form when primers hybridize to one another instead of the target template, competing for reaction resources and reducing the yield of your desired product [42].
2. What are the visual signs on a gel that my MgCl2 concentration is too high? If the MgCl2 concentration is too high, agarose gel electrophoresis of your PCR product may show:
- Multiple bands: Several bands of unexpected sizes, indicating nonspecific amplification [43].
- A smear: A fuzzy, continuous background of DNA, suggesting a multitude of nonspecific products [43].
- A strong, low molecular weight band: This is often a primer-dimer, typically appearing between 20-100 bp [42].
3. My PCR has failed. Could low MgCl2 be the cause? Yes. While high MgCl2 causes nonspecific binding, too little MgCl2 can lead to weak or no amplification. Magnesium is a critical cofactor for DNA polymerase; insufficient levels result in dramatically reduced enzyme activity [43]. The goal of optimization is to find the concentration that supports robust polymerase activity while maximizing primer-binding specificity.
Troubleshooting Guide: Stepwise MgCl2 Reduction Protocol
This protocol provides a systematic method to identify the optimal MgCl2 concentration for your specific reaction.
Objective: To eliminate nonspecific amplification and primer-dimers by identifying the minimum MgCl2 concentration that supports efficient amplification of your target.
Background Principle: The optimal concentration of MgCl2 is influenced by several factors in your reaction mix, including the concentration of dNTPs (which also bind Mg2+), the presence of EDTA from template preparation, and the specific primer-template combination [11] [44]. Therefore, optimization is required for each new set of primers.
Table 1: Reagents for MgCl2 Titration Experiment
| Reagent | Function | Notes for This Protocol |
|---|---|---|
| Template DNA | The DNA sequence to be amplified. | Use a high-quality, purified preparation. Typically 10 pg–1 µg per 50 µL reaction, depending on complexity [45] [44]. |
| Forward & Reverse Primers | Bind complementary regions to initiate amplification. | Optimize concentration (usually 0.1–1 µM). High concentrations promote primer-dimer formation [45] [11]. |
| DNA Polymerase | Enzyme that synthesizes new DNA strands. | Use a hot-start polymerase to minimize activity at low temperatures and reduce primer-dimer formation [42] [45] [11]. |
| dNTP Mix | Building blocks (dATP, dCTP, dGTP, dTTP) for new DNA. | Use balanced, equimolar concentrations. dNTPs chelate Mg2+, so their concentration directly affects free Mg2+ availability [11] [44]. |
| 10X PCR Buffer | Provides pH and salt conditions for optimal enzyme activity. | Often supplied Mg-free for optimization. Contains KCl to neutralize DNA charge and stabilize duplex formation [44]. |
| MgCl2 Stock Solution | Source of Mg2+ ions. | A 25 mM stock is commonly used. This is the variable reagent in this titration. |
Experimental Procedure:
- Prepare a Master Mix: Create a master mix containing all the common reagents for your number of reactions (plus ~10% extra to account for pipetting error): sterile water, 10X PCR buffer (without MgCl2), dNTPs, primers, DNA polymerase, and template DNA [46] [27].
- Aliquot the Master Mix: Dispense equal volumes of the master mix into 6-8 PCR tubes.
- Add MgCl2: Add MgCl2 from a stock solution to each tube to create a concentration gradient. A recommended starting range is 0.5 mM to 4.0 mM in increments of 0.5 mM [43].
- Run PCR: Place the tubes in a thermal cycler and run your standard PCR protocol.
- Analyze Results: Separate the PCR products by agarose gel electrophoresis. Identify the tube with the strongest desired band and the cleanest background (i.e., no nonspecific bands or primer-dimer).
Table 2: Expected Results from MgCl2 Titration
| MgCl2 Concentration | Expected Outcome | Recommended Action |
|---|---|---|
| Too Low (< 1.0 mM) | Faint or absent target band. | Gradually increase concentration. |
| Optimal (e.g., 1.5-2.5 mM) | Strong target band with a clean background. | Use this concentration. |
| Too High (> 3.0 mM) | Multiple bands, smearing, and/or strong primer-dimer band. | Gradually decrease concentration. |
The following workflow summarizes the logical process for diagnosing and correcting high MgCl2:
The Scientist's Toolkit: Essential Reagents for PCR Optimization
Table 3: Key Research Reagent Solutions
| Reagent | Function in PCR Optimization |
|---|---|
| Hot-Start DNA Polymerase | Polymerase that is inactive at room temperature, preventing nonspecific priming and primer-dimer formation during reaction setup. Requires high-temperature activation [45] [11] [27]. |
| MgCl2 Stock Solution (25 mM) | Allows for precise titration of magnesium ion concentration, which is critical for enzyme activity and reaction stringency [46] [43]. |
| PCR Additives (e.g., DMSO, Betaine) | Assist in amplifying difficult templates like GC-rich sequences by reducing secondary structures or increasing primer annealing stringency [43] [27] [44]. |
| dNTP Mix (balanced, 10 mM) | Provides the foundational nucleotides for DNA synthesis. Unbalanced dNTP concentrations can increase error rates and affect free Mg2+ levels [45] [11] [27]. |
Advanced Considerations and Integrated Optimization
Synergy with Annealing Temperature: The annealing temperature (Ta) and MgCl2 concentration are deeply interconnected. If you have optimized MgCl2 but still see minor nonspecific products, try increasing the annealing temperature in 1-2°C increments [45] [11]. A higher Ta increases stringency, requiring a more perfect match for primer binding, which can work synergistically with a corrected MgCl2 concentration to eliminate stubborn artifacts.
Mathematical Modeling for Prediction: Advanced research uses computational models to predict optimal PCR conditions. A recent study developed a predictive equation for MgCl2 concentration based on a multivariate Taylor series expansion, which achieved a high coefficient of determination (R² = 0.9942) [47]. The model highlights that the interaction between dNTP and primer concentrations is the most important variable (28.5% relative importance) for determining optimal MgCl2, followed by GC content (22.1%) and amplicon length (15.7%) [47]. This underscores why a one-size-fits-all MgCl2 concentration does not exist.
FAQ: How do I identify weak PCR amplification caused by low MgCl2?
Weak amplification due to insufficient MgCl₂ typically presents with specific symptoms in your results. On an agarose gel, you may observe either a very faint band of the correct expected size or no band at all [1]. This occurs because magnesium ions (Mg²⁺) are an essential cofactor for DNA polymerase; without a sufficient concentration, the enzyme's activity is drastically reduced, leading to inefficient primer extension and low product yield [1].
It is important to distinguish this from a complete PCR failure. If a negative control (a reaction with no template DNA) shows a similar faint smear or band, the issue is likely carry-over contamination rather than low MgCl₂ [16]. True low MgCl₂ failure will result in a clear, albeit faint or absent, target band with a clean background.
FAQ: What is the mechanism by which MgCl2 affects PCR yield?
Mg²⁺ plays two critical roles in the PCR reaction, both of which are compromised when its concentration is too low.
- Enzyme Cofactor: The Mg²⁺ ion is directly involved in the catalytic activity of DNA polymerase. It facilitates the formation of phosphodiester bonds by binding to a dNTP's alpha phosphate group, enabling the removal of beta and gamma phosphates and the subsequent attachment of the nucleotide to the growing DNA chain [1]. Without adequate Mg²⁺, this enzymatic process is inefficient or halts entirely.
- Nucleic Acid Stability: Mg²⁺ helps stabilize the binding of primers to the template DNA. It binds to the negatively charged phosphate backbone of the DNA, reducing the electrostatic repulsion between the primer and the template strand. This promotes more stable duplex formation and increases the primer's effective melting temperature (Tm), leading to more specific and efficient annealing [1].
The following diagram illustrates this core mechanism and its consequences when Mg²⁺ is low.
Troubleshooting Guide: Systematic Optimization of MgCl2 Concentration
Symptoms and Differentiating Factors
| Symptom on Agarose Gel | Potential Cause: Low MgCl₂ | Potential Cause: Other Issues (e.g., High MgCl₂) |
|---|---|---|
| Band Appearance | Faint or absent target band [1]. | Multiple non-specific bands or a smeared ladder [16] [5]. |
| Background | Clean background with no other products [1]. | Presence of primer-dimer or smearing [16] [5]. |
| Negative Control | Clean (no band) [16]. | May show smear or primer-dimer if issue is reagent contamination [16]. |
Optimization Experiment: MgCl2 Titration Protocol
To scientifically determine the optimal MgCl₂ concentration for your specific PCR assay, perform a titration experiment.
Detailed Methodology:
- Prepare a Master Mix: Create a master mix containing all standard PCR components—buffer (without Mg²⁺), DNA polymerase, dNTPs, primers, template DNA, and nuclease-free water. Omitting Mg²⁺ from the base buffer is crucial for an accurate titration [8].
- Aliquot the Master Mix: Dispense equal volumes of the master mix into 8 separate PCR tubes.
- Add MgCl₂: Add a calculated volume of 25 mM MgCl₂ stock solution to each tube to create a concentration gradient. The table below provides a standard scheme for a 50 µL reaction [16].
- Run the PCR: Place the tubes in a thermal cycler and start the optimized PCR program.
- Analyze the Results: Separate the PCR products by agarose gel electrophoresis. Identify the MgCl₂ concentration that produces the strongest, cleanest target band with the least background or non-specific product. This is the optimal concentration for your assay.
MgCl2 Titration Scheme for a 50 µL Reaction [16]:
| Tube | Final Mg²⁺ Concentration (mM) | Volume of 25 mM MgCl₂ Stock per 50 µL Reaction (µL) |
|---|---|---|
| 1 | 1.5 | 3.0 |
| 2 | 2.0 | 4.0 |
| 3 | 2.5 | 5.0 |
| 4 | 3.0 | 6.0 |
| 5 | 3.5 | 7.0 |
| 6 | 4.0 | 8.0 |
| 7 | 4.5 | 9.0 |
| 8 | 5.0 | 10.0 |
The Scientist's Toolkit: Essential Research Reagent Solutions
A successful PCR optimization relies on high-quality reagents. The following table details key materials and their functions.
| Reagent / Material | Function in PCR Optimization | Key Considerations |
|---|---|---|
| MgCl₂ Solution | Essential cofactor for DNA polymerase; stabilizes primer-template binding [1]. | Titrate between 1.5 - 5.0 mM; optimal is often 1.5 - 4.5 mM [16] [5] [1]. |
| DNA Polymerase | Enzyme that synthesizes new DNA strands. | Amount is critical; ~2.5 units/100 µL reaction. Excess can cause nonspecific products [16] [3]. |
| dNTP Mix | Building blocks (dATP, dCTP, dGTP, dTTP) for new DNA synthesis. | Use balanced, 200 µM of each dNTP. Excess can inhibit PCR [3] [8]. |
| Primers | Short oligonucleotides that define the start and end of the amplified sequence. | Typical final concentration is 0.1 - 1.0 µM. High concentrations cause mispriming [26] [16] [3]. |
| Template DNA | The DNA sample containing the target sequence to be amplified. | Quality is critical. Use 1 ng (plasmid) to 100 ng (genomic) DNA. Excess causes nonspecific amplification [26] [3]. |
| PCR Buffers | Provide optimal chemical environment (pH, ionic strength). | Often supplied with polymerase; may contain MgCl₂. Check composition before supplemental Mg²⁺ addition [8]. |
| Thermal Cycler | Instrument that automates temperature cycles for denaturation, annealing, and extension. | Ensure accurate temperature calibration and block uniformity for reproducible results. |
Experimental Workflow for Systematic PCR Optimization
The process of optimizing MgCl₂ to reduce nonspecific bands and boost yield is systematic. The following workflow diagrams the logical sequence of steps, from problem identification to a finalized, robust protocol.
FAQs: Core Concepts for Optimization
1. Why is it crucial to balance MgCl2 concentration, annealing temperature, and primer concentration in PCR?
These three factors are deeply interconnected in determining PCR specificity. MgCl2 acts as an essential cofactor for the DNA polymerase and stabilizes the binding between primers and the template DNA. This stabilization effectively increases the primer's melting temperature (Tm). If the MgCl2 concentration is increased without adjusting the annealing temperature, primers may bind to non-target sequences, leading to nonspecific bands. Conversely, a higher annealing temperature requires sufficient Mg2+ to facilitate stable primer-template binding. Primer concentration also plays a role, as high concentrations can exacerbate non-specific binding, especially when MgCl2 levels are suboptimal. Balancing all three is key to achieving clean, specific amplification [1] [13].
2. What are the direct consequences of getting the MgCl2 concentration wrong?
- Too much MgCl2 (e.g., >3-5 mM, depending on the reaction): Leads to decreased specificity by promoting non-specific primer binding and the appearance of extra, unwanted bands on a gel. It can also increase error rates by reducing the fidelity of the DNA polymerase [1] [11].
- Too little MgCl2 (e.g., <1 mM): Results in poor reaction efficiency, as the DNA polymerase enzyme is not sufficiently active. This can cause weak amplification or complete PCR failure due to primers failing to bind stably to the template [1].
3. How do I know if my PCR issues are due to MgCl2 and not something else?
Nonspecific amplification can have several causes. You should suspect MgCl2, annealing temperature, or primer issues if you observe the following on an agarose gel:
- Multiple bands of unexpected sizes [48] [11].
- A "smear" of DNA instead of sharp, discrete bands [48].
- Primer dimers, which appear as a bright band at the very bottom of the gel (typically 20-60 bp) [48]. Before concluding it is MgCl2, it is good practice to first verify the quality and quantity of your DNA template and the design of your primers [11] [49].
Troubleshooting Guides & Optimization Tables
Guide 1: Systematic Optimization of a New PCR Assay
Follow this workflow to establish robust conditions for your reaction, minimizing nonspecific amplification from the start.
Guide 2: Resolving Nonspecific Amplification
If you are already seeing nonspecific bands, use this targeted guide.
| Symptom | Possible Cause | Recommended Action |
|---|---|---|
| Multiple unexpected bands | Annealing temperature too low; Excessive MgCl2 | Increase annealing temperature in 1-2°C increments; Titrate down MgCl2 concentration [11] [13]. |
| Smear of DNA on gel | Excess MgCl2; Excess primers; Poor DNA quality | Titrate down MgCl2; Reduce primer concentration (e.g., to 0.2 µM); Check DNA integrity and purity [48] [11]. |
| Primer dimers (low molecular weight band) | Primer 3'-end complementarity; Low annealing temperature; Excess MgCl2 or primers | Redesign primers if possible; Increase annealing temperature; Optimize MgCl2 and primer concentrations [48] [11]. |
| Weak or no target band | MgCl2 concentration too low; Annealing temperature too high; Insufficient primers | Titrate up MgCl2 (e.g., to 2.0-3.0 mM); Lower annealing temperature; Ensure primer concentration is 0.1-1 µM [1] [49]. |
Table 1: Quantitative Optimization Ranges for Key Parameters
The following table summarizes evidence-based starting points and ranges for optimizing your reactions, synthesized from multiple studies.
| Parameter | Standard Starting Point | Optimal Range for Specificity | Special Considerations & Evidence |
|---|---|---|---|
| MgCl2 Concentration | 1.5 mM | 1.5 - 3.0 mM | A meta-analysis found a strong logarithmic relationship with DNA melting temperature. Each 0.5 mM increase within this range can raise Tm by ~1.2°C. Genomic DNA may require higher concentrations [15]. |
| Annealing Temperature | 5°C below primer Tm | 3 - 7°C below primer Tm (optimize via gradient) | For GC-rich templates, the optimal annealing temperature may be 7°C or more above the calculated Tm. Always use a gradient thermal cycler for empirical optimization [6]. |
| Primer Concentration | 0.5 µM | 0.1 - 1.0 µM | Concentrations as low as 0.2 µM can reduce non-specific product formation. High concentrations promote primer-dimer formation and nonspecific binding [13]. |
Table 2: Optimization for Challenging Templates (GC-Rich Sequences)
Amplifying GC-rich sequences often requires a synergistic adjustment of multiple parameters, as demonstrated in a study on the GC-rich EGFR promoter [6].
| Parameter | Standard Condition | Optimized Condition for GC-Rich Template |
|---|---|---|
| MgCl2 | 1.5 mM | 1.5 - 2.0 mM |
| Annealing Temperature | Calculated Tm | Calculated Tm + 7°C |
| Additive | None | 5% DMSO |
| DNA Template | Variable | At least 2 µg/mL |
The Scientist's Toolkit: Essential Research Reagents
This table details key reagents and their specific functions in optimizing PCR specificity.
| Reagent | Function in Optimization |
|---|---|
| MgCl2 | Essential cofactor for DNA polymerase; stabilizes primer-template binding and dNTP incorporation [1]. |
| Hot-Start DNA Polymerase | Reduces non-specific amplification and primer-dimer formation by remaining inactive until the first high-temperature denaturation step [11]. |
| PCR Additives (DMSO, BSA, Betaine) | Help denature GC-rich templates and secondary structures, improving specificity and yield. DMSO was crucial for amplifying the GC-rich EGFR promoter [8] [6]. |
| dNTP Mix (equimolar) | Unbalanced dNTP concentrations can increase PCR error rates and inhibit the reaction. Mg2+ binds to dNTPs, so their concentration can affect free Mg2+ availability [11] [13]. |
| Gradient Thermal Cycler | Allows empirical determination of the optimal annealing temperature by testing a range of temperatures simultaneously in a single run [11]. |
FAQs on Nonspecific Bands in PCR
What causes multiple nonspecific bands to appear on my agarose gel? Multiple nonspecific bands indicate that your primers are binding to and amplifying unintended regions of the template DNA. Common causes include suboptimal annealing temperature, excessive magnesium chloride (MgCl₂) concentration, poor primer design, or too much primer or template DNA in the reaction [13] [10].
Why is MgCl₂ concentration so critical for reaction specificity? Magnesium ions (Mg²⁺) are an essential cofactor for DNA polymerase activity, but they also stabilize the DNA duplex. Too low a concentration results in weak or no amplification, while too high a concentration reduces primer annealing stringency, making it easier for primers to bind to non-target sequences and produce nonspecific bands [13] [50] [51]. A recent meta-analysis established a clear logarithmic relationship between MgCl₂ concentration and DNA melting temperature, highlighting the need for precise concentration control [15].
My PCR used to work perfectly. Why am I now seeing smears and nonspecific bands? This is a common problem often caused by the gradual accumulation of "amplifiable DNA contaminants" in the lab environment that interact with your specific primers. The most efficient solution is to switch to a new set of primers with different sequences. Other measures include separating pre- and post-PCR workspaces and using fresh reagent aliquots [10] [52].
What is a "primer dimer," and how is it different from nonspecific bands? A primer dimer is a specific type of nonspecific amplification product formed when two primers hybridize to each other and get extended by the polymerase. It typically appears as a very bright band at the bottom of the gel (around 20-60 bp), much smaller than your target band. In contrast, other nonspecific bands can be of various sizes, including larger than your target [48] [8].
Troubleshooting Guide: A Systematic Approach to Resolving Multiple Nonspecific Bands
Follow this logical workflow to diagnose and fix the issue of multiple nonspecific bands.
Step 1: Investigate and Optimize Primer Design
Poor primer design is a frequent culprit. Re-evaluate your primers using these criteria [8] [27]:
- Length: 15-30 nucleotides.
- GC Content: 40-60%.
- Melting Temperature (Tm): 52-58°C for both primers, with a difference of no more than 5°C.
- 3' End: Avoid complementarity between the 3' ends of the forward and reverse primers to prevent primer-dimer formation [8].
- Specificity: Use tools like NCBI Primer-BLAST to verify target specificity.
Step 2: Increase the Annealing Temperature
The annealing temperature (Ta) is critical for specificity. If the Ta is too low, primers can bind imperfectly to non-target sites.
- Action: Increase the annealing temperature incrementally by 1-2°C [53]. Using a thermal cycler with a gradient function is the most efficient way to find the optimal Ta [51]. A higher Ta promotes more specific primer binding [13].
Step 3: Titrate Magnesium Chloride (MgCl₂) Concentration
As a core part of a thesis on MgCl₂ optimization, systematically calibrating its concentration is essential. Mg²⁺ concentration directly influences the stability of primer-template binding [13] [51].
- Protocol for MgCl₂ Titration:
- Prepare a master mix without MgCl₂ or with a known baseline concentration.
- Set up a series of reactions with MgCl₂ concentrations ranging from 1.0 mM to 4.0 mM in 0.5 mM increments [50] [51].
- Run the PCR and analyze the products by gel electrophoresis.
- The optimal concentration is the one that yields a single, bright band of the expected size with the least background or nonspecific bands [50].
Step 4: Adjust Reaction Component Volumes
- Primer Concentration: High primer concentrations can promote mispriming and primer-dimer formation. Test concentrations between 0.1 μM and 0.5 μM [13] [27].
- Template Quantity: Too much template DNA can introduce nonspecific binding sites and cause smearing. Reduce the template amount, especially if the DNA is of high concentration [52].
Step 5: Utilize High-Stringency Techniques and Additives
- Hot-Start PCR: Use a hot-start DNA polymerase. These enzymes are inactive until the initial high-temperature denaturation step, preventing enzymatic activity during reaction setup and reducing non-specific amplification at lower temperatures [10] [27].
- PCR Additives: For particularly stubborn cases, especially with GC-rich templates, additives can be highly effective.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Optimizing PCR Specificity
| Reagent | Function in PCR | Optimization Guidance |
|---|---|---|
| MgCl₂ Solution | Essential cofactor for DNA polymerase; stabilizes primer-template binding [50] [51]. | Titrate between 1.0-4.0 mM. The optimal concentration is often between 1.5-3.0 mM [15] [50]. |
| Hot-Start DNA Polymerase | Remains inactive at room temperature, preventing non-specific priming during reaction setup [10]. | Prefer over standard polymerase. Ideal for high-specificity applications. |
| dNTP Mix | Building blocks for new DNA strands. | Use balanced concentrations of dATP, dCTP, dGTP, and dTTP. High or degraded dNTPs can increase error rates [53]. |
| PCR Additives (e.g., DMSO) | Modifies nucleic acid melting behavior and reduces secondary structures, improving specificity and yield for difficult templates [51] [27]. | Test at recommended concentrations (e.g., 1-10% for DMSO). |
| Thermostable Polymerase Buffer | Provides the optimal ionic environment (pH, salts) for polymerase activity and fidelity. | Always use the buffer supplied by the enzyme manufacturer, as it is formulated for optimal performance. |
Beyond Trial and Error: Validating Conditions with Predictive Models and Quantitative Analysis
In the polymerase chain reaction (PCR), magnesium chloride (MgCl₂) is a critical cofactor that directly influences the enzyme activity of DNA polymerase and the fidelity of the entire amplification process [1]. Its concentration is a key determinant in achieving a balance between high yield and specific amplification of the intended target. Incorrect MgCl₂ levels are a primary cause of nonspecific amplification, leading to ambiguous results and failed experiments. These guidelines synthesize recent meta-analysis findings to provide evidence-based protocols for optimizing MgCl₂ concentration, with the specific aim of reducing nonspecific PCR bands.
Core Quantitative Findings from Meta-Analysis
A systematic meta-analysis of 61 peer-reviewed studies provides robust, quantitative data on the relationship between MgCl₂ concentration and PCR performance [54] [15]. The findings offer a solid foundation for moving beyond empirical optimization.
Table 1: Evidence-Based MgCl₂ Guidelines from Meta-Analysis
| Parameter | Optimal Range | Quantitative Effect | Key Influencing Factor |
|---|---|---|---|
| Overall Optimal MgCl₂ Range | 1.5 - 3.0 mM | N/A | General standard PCR [54] [15] |
| DNA Melting Temperature (Tm) | N/A | Increases by ~1.2°C per 0.5 mM MgCl₂ | Logarithmic relationship within 1.5-3.0 mM range [54] |
| Template-Specific Requirements | Genomic DNA: Higher end of rangeSimple Templates: Lower end of range | Concentration must be tailored | Template complexity and GC content [54] |
These findings establish that MgCl₂ concentration is not a one-size-fits-all parameter. Precise modulation, tailored to the specific template and primer system, is essential for achieving both high efficiency and specificity [54].
Troubleshooting Guide: Resolving Nonspecific Amplification
The following section addresses common issues related to MgCl₂ concentration in a question-and-answer format.
FAQ 1: What happens if I add too much MgCl₂ to my PCR reaction?
- Answer: Excessive MgCl₂ (typically >3.0 mM, though this can vary) has several detrimental effects [5] [1]:
- Increased Non-Specific Binding: High Mg²⁺ concentration stabilizes DNA duplexes non-specifically, allowing primers to bind to incorrect, partially complementary sites on the template DNA. This results in multiple bands or a smeared background on an agarose gel [13] [5] [1].
- Formation of Primer Dimers: The elevated ionic conditions facilitate the annealing of primers to each other, creating short, artifactual "primer dimer" products that compete with the amplification of the target DNA [13] [5].
- Reduced Stringency: The reaction becomes more tolerant of mismatches between the primer and template, drastically reducing specificity [13].
FAQ 2: What are the symptoms of too little MgCl₂, and how do they differ from too much?
- Answer: Insufficient MgCl₂ presents a different set of problems [1]:
- Weak or No Amplification: Mg²⁺ is an essential cofactor for Taq DNA polymerase. At low concentrations, enzyme activity is significantly reduced, leading to very low yield or complete PCR failure [1].
- A Single, Faint, or Absent Band: On a gel, the result is the opposite of a high Mg²⁺ reaction. Instead of multiple bands, you will see little to no product [1].
- Key Difference: While high MgCl₂ causes "too many" bands, low MgCl₂ causes "too few" or no bands. This distinction is the first step in effective troubleshooting.
FAQ 3: How does template DNA type influence the optimal MgCl₂ concentration?
- Answer: The meta-analysis confirmed that template complexity directly affects Mg²⁺ requirements [54].
- Genomic DNA: Requires higher concentrations of MgCl₂ (often towards the 3.0 mM end of the optimal range or slightly higher) due to its complexity and the potential for polymerase inhibitors in the sample that may chelate Mg²⁺ ions [54].
- Plasmid or Simple Templates: Can be efficiently amplified with lower concentrations of MgCl₂ (closer to 1.5 mM), as there are fewer non-specific sites for primer binding [54].
- GC-Rich Templates: These challenging sequences often require specialized optimization, sometimes involving higher MgCl₂ and specific additives [55].
Experimental Protocol: Optimizing MgCl₂ Concentration
This protocol provides a step-by-step methodology for empirically determining the optimal MgCl₂ concentration for your specific PCR assay.
Objective: To identify the MgCl₂ concentration that produces a strong, single band corresponding to your target amplicon, with minimal to no non-specific background.
Required Reagents:
- Table 2: Research Reagent Solutions for MgCl₂ Optimization
| Reagent | Function in the Experiment |
|---|---|
| 10X PCR Buffer (without MgCl₂) | Provides the core reaction environment (pH, salts). Using a Mg-free buffer is essential for this test. |
| MgCl₂ Stock Solution (e.g., 25 mM) | The variable being tested to establish its optimal final concentration. |
| dNTP Mix | The building blocks for DNA synthesis. |
| Forward and Reverse Primers | Designed to specifically anneal to the target sequence. |
| DNA Polymerase (e.g., Taq) | The enzyme that catalyzes DNA synthesis; requires Mg²⁺ as a cofactor. |
| Template DNA | The sample containing the target sequence to be amplified. |
| Agarose Gel Electrophoresis System | Used to separate and visualize the PCR products to assess specificity and yield. |
Detailed Methodology:
Prepare a Master Mixture: In a sterile 1.5 ml microcentrifuge tube, combine all the common PCR components for the number of reactions in your test gradient plus one extra to account for pipetting error. This includes sterile water, PCR buffer (without MgCl₂), dNTPs, primers, template DNA, and DNA polymerase [8]. Mix thoroughly by pipetting up and down gently.
Aliquot and Vary MgCl₂: Pipette equal volumes of the master mixture into individual PCR tubes. Then, add a different volume of MgCl₂ stock solution to each tube to create a final concentration gradient. A typical and effective range is from 1.0 mM to 4.0 mM in 0.5 mM increments (e.g., 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0 mM) [55]. Include a negative control (no template DNA) for one of the middle concentrations.
Run PCR: Place the tubes in a thermal cycler and start the PCR program using your standard cycling conditions. If non-specificity is a persistent issue, consider pairing this MgCl₂ gradient with a slightly elevated annealing temperature.
Analyze Results: Separate the PCR products using agarose gel electrophoresis. Visualize the DNA bands under UV light and document the results.
Interpretation and Identification:
- No/Low Yield Tubes (Low [MgCl₂]): These reactions will show a faint or absent target band.
- Optimal Tubes: These will show a single, bright band of the expected size.
- Non-Specific Tubes (High [MgCl₂]): These will show multiple bands, smearing, or primer dimers. The goal is to identify the concentration that produces the strongest specific signal with the cleanest background. This is your optimal MgCl₂ concentration.
The Scientist's Toolkit: Advanced Scenarios & Reagents
For standard PCR, the protocol above is sufficient. However, specific challenges require advanced tools.
GC-Rich Amplification: GC-rich templates (>60% GC content) are problematic due to stable secondary structures and higher melting temperatures [55].
- Strategy: In addition to testing a MgCl₂ gradient, incorporate specialized additives into your optimization.
- Key Reagents:
- Betaine: Can be used at a final concentration of 0.5 M to 2.5 M. It disrupts base stacking, effectively equalizing the stability of GC and AT pairs and facilitating denaturation [8].
- DMSO: Used at 1-10%, it helps denature DNA secondary structures and can improve primer specificity [55] [8].
- GC-Enhanced Polymerases: Commercial polymerases like Q5 or OneTaq are often supplied with proprietary GC enhancers that combine multiple additives for optimal performance with difficult templates [55].
Preventing Genotyping Errors: Incorrect MgCl₂ concentration can lead to allele-dependent amplification biases, skewing genotype results and violating Hardy-Weinberg equilibrium [56].
- Strategy: Meticulously optimize MgCl₂ concentration for any genotyping assay. The meta-analysis confirms that precise Mg²⁺ modulation is crucial for reliable and reproducible results, especially in diagnostic and genetic studies [54] [56].
Optimizing MgCl₂ is a fundamental and non-negotiable step in developing a robust, specific PCR assay. The evidence-based optimal range of 1.5 to 3.0 mM serves as a critical starting point [54] [15]. However, the key to eliminating nonspecific bands lies in the systematic, empirical determination of the perfect concentration for your unique primer-template system. By following the structured troubleshooting guide and experimental protocol outlined in this document, researchers can confidently overcome the challenge of nonspecific amplification, ensuring the accuracy and reliability of their molecular data.
FAQ: Troubleshooting Nonspecific PCR Amplification
What are the primary causes of nonspecific PCR bands, and how can I resolve them?
Nonspecific amplification, visible as multiple bands or smears on a gel, is a common issue in PCR. The table below summarizes the primary causes and their solutions.
| Primary Cause | Specific Issue | Recommended Solution |
|---|---|---|
| Reaction Components | Excess Mg2+ concentration | Optimize Mg2+ concentration in 0.2–1 mM increments; high concentrations favor mispriming [11] [57]. |
| Inappropriate or excess DNA polymerase | Use hot-start DNA polymerases to prevent activity at room temperature; review and decrease enzyme amount if necessary [11] [57]. | |
| Incorrect primer concentration | Optimize primer concentrations (typically 0.1–1 µM); high concentrations promote primer-dimer formation [11] [57]. | |
| Thermal Cycling | Annealing temperature too low | Increase the annealing temperature; the optimal is usually 3–5°C below the lowest primer Tm. Use a gradient cycler for optimization [11] [8]. |
| Excessive number of cycles | Reduce the number of cycles (generally to 25–35) to prevent accumulation of nonspecific amplicons [11] [16]. | |
| Insufficient denaturation | Increase denaturation time and/or temperature for GC-rich templates or sequences with secondary structures [11]. | |
| Template & Primers | Poor primer design | Verify primers are specific to the target, have no self-complementarity, and have similar Tm values (difference <5°C) [8] [57]. |
| Excess template DNA | Review and lower the quantity of input DNA to reduce nonspecific products [11] [16]. |
How does MgCl2 concentration specifically influence the formation of nonspecific bands?
Magnesium ions (Mg2+) are a critical cofactor for DNA polymerase activity. However, deviation from the optimal concentration is a major contributor to nonspecific amplification.
- Mechanism of Effect: Mg2+ stabilizes the double-stranded DNA and the primer-template complex. An excessively high concentration reduces the stringency of primer annealing, allowing primers to bind to non-target sequences with partial complementarity, leading to nonspecific amplification [11] [57].
- Quantitative Optimization Range: The optimal Mg2+ concentration is typically between 1.5 mM and 5.0 mM [16]. The following table provides a precise pipetting guide for optimization using a standard 25 mM MgCl2 stock solution in a 50 µl reaction [16].
| Desired Final [Mg2+] (mM) | 1.5 | 2.0 | 2.5 | 3.0 | 3.5 | 4.0 | 4.5 | 5.0 |
|---|---|---|---|---|---|---|---|---|
| Volume of 25 mM MgCl2 per 50 µl reaction (µl) | 0 | 2 | 4 | 6 | 8 | 10 | 12 | 14 |
Can machine learning assist in PCR optimization to prevent such issues?
Yes, machine learning (ML) is an emerging powerful tool for predictive PCR assay design and optimization. ML models can analyze sequence features to predict the success of amplification, thereby reducing reliance on empirical troubleshooting.
- Principle: ML models are trained on large datasets from past PCR experiments. They learn to identify key features—such as primer mismatch patterns, melting temperature (Tm), and GC content—that correlate with amplification efficiency and specificity [58].
- Application Example: The BioInnovate AI platform uses a Light Gradient Boosting Machine (LGBM) model to predict effective PCR primer-probe combinations. This platform achieved an Area Under the Curve (AUC) of 0.97, significantly reducing assay development time by approximately 90% [58]. By predicting optimal reagents and conditions upfront, these frameworks can preemptively circumvent common causes of failure, including nonspecific amplification.
Experimental Protocol: Systematic Optimization of MgCl2 Concentration
Objective
To empirically determine the optimal MgCl2 concentration for a specific PCR assay to maximize target yield and minimize nonspecific amplification.
Materials and Reagents
| Research Reagent Solution | Function in the Experiment |
|---|---|
| Template DNA (e.g., genomic DNA) | The target DNA sequence to be amplified. |
| Forward and Reverse Primers | Short, single-stranded DNA sequences that define the region to be amplified. |
| 10X PCR Buffer (often supplied Mg-free) | Provides the optimal chemical environment (pH, salts) for the DNA polymerase. |
| 25 mM MgCl2 Stock Solution | The variable cofactor being optimized; essential for DNA polymerase activity. |
| dNTP Mix (e.g., 10 mM) | The building blocks (dATP, dCTP, dGTP, dTTP) for synthesizing new DNA strands. |
| DNA Polymerase (e.g., Taq) | The enzyme that catalyzes the synthesis of new DNA strands. |
| Sterile Nuclease-Free Water | Used to bring the reaction to the final volume. |
Methodology
- Preparation: Thaw all PCR reagents completely on ice. Prepare a Master Mix for all common components to minimize pipetting error and ensure consistency across reactions.
- Master Mix Composition (for a single 50 µl reaction): Combine the following in a sterile tube:
- 5.0 µl of 10X PCR Buffer (Mg-free)
- 1.0 µl of 10 mM dNTP Mix
- 1.0 µl of 20 µM Forward Primer
- 1.0 µl of 20 µM Reverse Primer
- 0.5 µl of DNA Polymerase (e.g., 0.5 Units/µl)
- X µl of Template DNA (e.g., 1-1000 ng)
- Y µl of Nuclease-Free Water (The volume of water 'Y' will depend on the volume of MgCl2 added in the next step to keep the final volume at 50 µl) [8].
- Aliquoting and MgCl2 Titration:
- Aliquot the appropriate volume of the Master Mix into eight separate 0.2 ml PCR tubes.
- To each tube, add the volume of 25 mM MgCl2 stock solution as specified in the quantitative table above (e.g., 0 µl, 2 µl, 4 µl, ..., 14 µl) [16].
- Gently mix the reactions by pipetting up and down.
- Thermal Cycling: Place the tubes in a thermal cycler and run the standard PCR protocol optimized for your primer pair and template.
- Analysis: Analyze the PCR products using agarose gel electrophoresis. The lane with the strongest specific band and the least background smear or nonspecific bands indicates the optimal MgCl2 concentration for your assay.
Framework Integration: From Thermodynamic Principles to Machine Learning Prediction
The process of optimizing a PCR reaction, such as MgCl2 concentration, is fundamentally guided by the thermodynamics of nucleic acid interactions. Machine learning frameworks build upon this by quantifying and modeling these relationships to make accurate predictions.
Workflow Diagram
Key Feature Analysis for Predictive Modeling
Machine learning models, such as the one used in the BioInnovate AI platform, rely on specific, quantifiable features derived from thermodynamic principles to predict PCR success. The most influential features identified through analyses like SHAP (Shapley Additive Explanations) include [58]:
- Total Mismatches (total_mm): The overall number of base pair mismatches between the primer and the target template. A higher count strongly correlates with amplification failure.
- 3'-End Mismatch Percentage (3pmmpercent): The proportion of mismatches within the first five base pairs from the 3' end of the primer. This is critically important for priming efficiency.
- Terminal Mismatch Percentage (termmmpercent): The presence of a mismatch at the very last (3' terminal) base of the primer, which drastically reduces amplification efficiency.
- Melting Temperature (Tmmean) and Variation (Tmdiff): The average Tm of the primer pair and the difference between their individual Tm values. A large difference can lead to inefficient amplification.
By integrating these thermodynamic parameters into a predictive ML model, researchers can move from a reactive troubleshooting approach to a proactive, optimized assay design, thereby enhancing the reliability and speed of molecular diagnostics and research [58].
Troubleshooting FAQs: MgCl2 Optimization
FAQ 1: What is the fundamental role of MgCl2 in a PCR reaction, and why is its concentration so critical?
MgCl2 is an essential cofactor for DNA polymerase activity. Magnesium ions (Mg2+) directly enable the enzyme to incorporate dNTPs by catalyzing the formation of phosphodiester bonds during polymerization [3]. Furthermore, Mg2+ stabilizes the DNA double helix by binding to the negatively charged phosphate backbone of both DNA templates and primers. This binding reduces electrostatic repulsion, facilitating primer annealing and stabilizing the primer-template complex [4] [3]. Its concentration is vital because it directly affects nearly every aspect of PCR thermodynamics, including DNA melting temperature, primer annealing efficiency, and reaction specificity [4] [13]. An incorrect concentration is a primary cause of PCR failure, leading to problems like the complete absence of a product, the appearance of nonspecific bands, or the formation of primer-dimers [8] [13].
FAQ 2: I consistently observe nonspecific bands or a smeared background on my agarose gel. How can adjusting MgCl2 help resolve this?
Nonspecific amplification and smearing often indicate that the primer annealing is not sufficiently specific, which can be caused by excessively high MgCl2 concentration. Elevated Mg2+ levels over-stabilize the primer-template interaction, allowing primers to bind to non-target sequences with partial complementarity [4] [13]. To troubleshoot this:
- Decrease the Concentration: Systematically lower the MgCl2 concentration in 0.5 mM increments from your starting point. A reduction can increase reaction stringency, favoring only the perfect primer-template matches [13].
- Find the Optimal Range: The meta-analysis identifies 1.5–3.0 mM as a common optimal range for efficient performance, but the ideal value is template-dependent [4]. A typical starting concentration is 1.5 mM, but optimization from 0.5 mM to 5.0 mM is sometimes necessary [8] [59].
FAQ 3: My PCR reaction yielded no product. Should I increase the MgCl2 concentration?
A failed amplification could be due to insufficient Mg2+ for the DNA polymerase to function or for stable primer-template complexes to form [4] [3]. Before adjusting MgCl2, confirm other factors like template quality and integrity. If these are satisfactory, consider increasing MgCl2. A gradual increase of 0.5 mM per test reaction can help determine the optimal level. Remember that the required Mg2+ concentration is also influenced by the concentration of dNTPs, as Mg2+ binds to dNTPs in the reaction. If you increase dNTPs, you may also need to increase MgCl2 to ensure a sufficient amount of free Mg2+ ions remains available for the polymerase [3].
Table 1: Key Quantitative Relationships for MgCl2 in PCR
| Parameter | Relationship / Optimal Range | Key Findings from Meta-Analysis & Studies |
|---|---|---|
| General Optimal Range | 1.5 - 4.5 mM [59] | A broad starting point for many standard PCR applications. |
| Theoretical Optimal Range | 1.5 - 3.0 mM [4] | Identified via meta-analysis as optimal for efficient PCR performance. |
| Effect on DNA Melting Temperature (Tm) | +1.2 °C per 0.5 mM MgCl2 [4] | A strong logarithmic relationship exists; increasing MgCl2 raises the Tm. |
| Template-Specific Guidance | Higher for complex templates [4] | Genomic DNA templates often require higher MgCl2 than simple plasmid DNA. |
Table 2: Predictive Model for MgCl2 Concentration (from Mathematical Modeling)
The following equation was developed using multivariate Taylor series expansion and thermodynamic principles to predict optimal MgCl2 concentration, achieving an R² of 0.9942 [47]: (MgCl2) ≈ 1.5625 + (-0.0073 × Tm) + (-0.0629 × GC) + (0.0273 × L) + (0.0013 × dNTP) + (-0.0120 × Primers) + (0.0007 × Polymerase) + (0.0012 × log(L)) + (0.0016 × TmGC) + (0.0639 × dNTPPrimers) + (0.0056 × pH_Polymerase) [47]
| Variable | Description | Relative Importance in Model |
|---|---|---|
| dNTP_Primers | Interaction between dNTP and primer concentrations | 28.5% |
| GC | GC content of the template (%) | 22.1% |
| L | Amplicon length (base pairs) | 15.7% |
| Tm | Primer melting temperature (°C) | 12.3% |
Detailed Experimental Protocol: MgCl2 Concentration Gradient
This protocol provides a methodology for empirically determining the optimal MgCl2 concentration for any specific PCR reaction.
Objective: To identify the MgCl2 concentration that yields the highest specificity and efficiency for a given primer-template combination.
The Scientist's Toolkit: Research Reagent Solutions
| Reagent | Function & Optimization Notes | Typical 50 µL Reaction [8] |
|---|---|---|
| Template DNA | The DNA target to be amplified. Complexity affects MgCl2 needs; genomic DNA often requires more than plasmid DNA [4] [3]. | 1-1000 ng (e.g., 0.5 µl of 2 ng/µl) |
| Taq DNA Polymerase | Enzyme that synthesizes new DNA strands. Its activity is strictly Mg2+-dependent [60] [3]. | 0.5-2.5 units (e.g., 0.5 µl of 0.5 U/µl) |
| 10X PCR Buffer | Provides pH and salt conditions for optimal enzyme activity. Often supplied with or without MgCl2 [8] [60]. | 5 µl |
| Primers (Forward & Reverse) | Short DNA sequences that define the region to be amplified. Concentration and design are critical for specificity [8] [3]. | 20-50 pmol each (e.g., 1 µl of 20 µM each) |
| dNTP Mix | Building blocks (dATP, dCTP, dGTP, dTTP) for new DNA strands. Compete with primers for Mg2+ binding [3]. | 200 µM total (e.g., 1 µl of 10 mM mix) |
| MgCl2 Solution | Critical variable. Cofactor for polymerase and stabilizer of nucleic acid interactions [4] [3]. | Variable (e.g., 0-10 µl of 25 mM stock) |
| Sterile Water | Nuclease-free water to bring the reaction to the final volume. | Q.S. to 50 µl |
Procedure:
- Prepare a Master Mix: Calculate the reagents needed for ( n+1 ) reactions (where ( n ) is the number of MgCl2 conditions to test, plus one for the negative control). In a sterile 1.5 ml microcentrifuge tube, combine the following components on ice: sterile water, 10X PCR buffer (without MgCl2), dNTP mix, forward primer, reverse primer, and DNA polymerase. Mix gently by pipetting up and down [8].
- Aliquot the Master Mix: Dispense equal volumes of the Master Mix into each PCR tube.
- Spike in MgCl2: Add a varying volume of MgCl2 stock solution (e.g., 25 mM) to each tube to create a concentration gradient. For example, prepare tubes to achieve final concentrations of 0.5, 1.0, 1.5, 2.0, 3.0, 4.0, and 5.0 mM. One tube should serve as a negative control (no template DNA) [8] [60].
- Add Template DNA: Add the template DNA to each reaction tube, except for the negative control.
- Thermal Cycling: Place the tubes in a thermal cycler and run the appropriate program. A typical program includes:
- Initial Denaturation: 94-95°C for 2-5 minutes.
- Amplification (25-35 cycles):
- Final Extension: 72°C for 5-10 minutes.
- Hold: 4-10°C indefinitely.
- Analyze Results: Separate the PCR products by agarose gel electrophoresis. Analyze the gel for the presence of a single, sharp band of the expected size at the various MgCl2 concentrations. The condition that produces the brightest specific band with the least background smearing or nonspecific bands is the optimal MgCl2 concentration [8].
Workflow and Relationship Visualization
MgCl2 Troubleshooting Logic
MgCl2 Concentration Impact on PCR
In the context of research focused on reducing nonspecific PCR bands by optimizing MgCl₂ concentration, rigorous validation of your assays is not optional—it is fundamental to generating reliable, reproducible data. The powerful amplification capability of PCR and quantitative PCR (qPCR) means that even minor deviations in reaction components or conditions can lead to misleading results, such as false positives, false negatives, or inaccurate quantification. These issues are often visible as smeared bands, multiple bands, or faint bands in gel electrophoresis, or they can manifest as poor amplification efficiency and specificity in qPCR. Adhering to established validation guidelines ensures that your optimized MgCl₂ conditions truly enhance assay performance rather than introducing new variables or artifacts. This guide provides targeted troubleshooting and methodological support to help you confirm that your assays are both specific and efficient.
Troubleshooting Guide: FAQs for Common Experimental Issues
This section addresses specific problems you might encounter when validating PCR and qPCR assays.
FAQ 1: My agarose gel shows smeared bands. What could be the cause and how can I fix it?
Smeared bands are a common issue that can stem from several factors related to sample quality, reaction conditions, and gel procedure [61] [62].
- Possible Cause: Excessive Template DNA. Overloading the reaction with too much template DNA is a frequent cause of smearing [62].
- Solution: Reduce the amount of template DNA in your reaction. A general recommendation is to load 0.1–0.2 μg of DNA per millimeter of gel well width [61].
- Possible Cause: Degraded DNA Template. Degraded DNA will appear as a smear of fragments of various sizes [61] [11].
- Solution: Re-isolate your DNA template using good laboratory practices to prevent nuclease contamination. Verify DNA integrity by running a sample on a gel before PCR [62] [11].
- Possible Cause: Suboptimal PCR Conditions. Low annealing temperatures or long extension times can lead to non-specific amplification and smearing [10] [62].
- Solution: Increase the annealing temperature stepwise to improve specificity. You can also reduce the extension time [62]. For a more systematic approach, use a thermal cycler with a gradient function to optimize these parameters.
- Possible Cause: Contamination. The gradual accumulation of amplifiable DNA contaminants can cause smearing, even with previously reliable primers [10].
- Solution: Implement strict laboratory practices to separate pre- and post-PCR workspaces. If contamination is suspected, a highly effective solution is to switch to a new set of primers with different sequences [10].
FAQ 2: I am getting faint or no bands on my gel after PCR. What should I check?
This problem typically indicates a failure of the amplification reaction itself, often due to insufficient reaction components or inactive reagents [62].
- Possible Cause: Low Quantity or Purity of DNA Template. The concentration of your template might be too low, or it might be contaminated with inhibitors like phenol or salts [10] [11].
- Solution: Check the concentration and purity of your DNA template (a 260/280 nm ratio of ~1.8 is ideal for DNA). If necessary, purify or concentrate the template. Increase the number of PCR cycles (e.g., to 35-40) if the starting template is very low [62] [11].
- Possible Cause: Insufficient Reagents. Key reaction components may be missing, degraded, or at too low a concentration.
- Solution: Use fresh aliquots of all reagents, especially dNTPs and polymerase. Ensure your primer concentration is adequate (typically 0.1–1 μM) and consider increasing it slightly [62] [11]. Verify that Mg²⁺ concentration is sufficient, as it is a critical cofactor for polymerase activity [10] [11].
- Possible Cause: Incorrect Thermal Cycler Programming. The denaturation, annealing, or extension steps may not be set correctly.
- Solution: Double-check your PCR program. Ensure the denaturation temperature is high enough (typically 94-98°C) and that the annealing temperature is appropriate for your primers [11].
FAQ 3: My qPCR results show poor amplification efficiency or non-specific signals. How can I improve validation?
qPCR validation requires careful attention to primer design and reaction linearity to ensure accurate quantification [63] [64].
- Possible Cause: Poor Primer Design or Quality. Primers with low specificity can form primer-dimers or amplify non-target sequences, reducing efficiency [65].
- Solution: Redesign primers using specialized software to ensure specificity and optimal melting temperature (Tm). Check for primer-dimer formation with a melt curve analysis [65].
- Possible Cause: Assay Not Validated for Linear Dynamic Range. Quantitative results are only reliable within a defined range of template concentrations [64].
- Solution: Perform a dilution series with a known standard. The assay's linear dynamic range is established by creating a standard curve from a seven 10-fold dilution series run in triplicate. A correlation coefficient (R²) of ≥ 0.980 and a primer efficiency between 90% and 110% are generally considered acceptable [64].
- Possible Cause: Lack of Inclusivity/Exclusivity Testing. The assay may not reliably detect all intended targets (inclusivity) or may cross-react with non-targets (exclusivity) [64].
- Solution: Validate both in silico (using sequence databases) and experimentally at the bench. Test your assay against a panel of well-defined target and non-target strains to confirm its accuracy [64].
Key Experimental Protocols for Validation
Protocol: Determining the Optimal MgCl₂ Concentration
Optimizing Mg²⁺ is crucial as it acts as a cofactor for DNA polymerase and affects primer annealing [66] [11]. This protocol is central to a thesis on reducing nonspecific bands.
- Prepare a Master Mix: Create a master mix containing all PCR components except MgCl₂ and template DNA.
- Set Up a Gradient: Aliquot the master mix into several tubes. Add MgCl₂ to each tube to create a concentration gradient. A typical range to test is 1.0 mM to 4.0 mM in 0.5 mM increments [66].
- Run PCR: Add template DNA to each tube and run the PCR under your standard cycling conditions.
- Analyze Results: Analyze the PCR products using gel electrophoresis. Identify the Mg²⁺ concentration that yields the strongest specific band with the least background smearing or non-specific bands. This is your optimal concentration.
Protocol: Validating qPCR Amplification Efficiency and Dynamic Range
This protocol is essential for confirming that your qPCR assay produces accurate, quantitative data [64].
- Prepare a Standard Curve: Serially dilute a sample of known concentration (e.g., a synthetic oligonucleotide or a pre-quantified cDNA) in a seven-step, 10-fold dilution series.
- Run qPCR: Amplify each dilution in triplicate using your qPCR assay.
- Analyze the Curve: Plot the log of the starting template quantity against the Ct value obtained for each dilution.
- Calculate Efficiency: The slope of the resulting standard curve is used to calculate amplification efficiency using the formula: Efficiency = [10^(-1/slope) - 1] x 100%. An efficiency of 90-110% is optimal.
- Assess Linearity: The correlation coefficient (R²) of the standard curve should be ≥ 0.980 to demonstrate a strong linear relationship [64].
Workflow: Systematic Troubleshooting for Gel Electrophoresis Problems
The following diagram outlines a logical workflow for diagnosing and resolving common gel electrophoresis issues, connecting observable problems to their root causes and solutions.
The Scientist's Toolkit: Essential Research Reagents and Materials
The following table details key reagents and materials critical for successfully validating PCR and qPCR assays, especially in the context of optimizing parameters like MgCl₂.
| Item | Function / Explanation in Validation |
|---|---|
| High-Fidelity or Hot-Start Polymerase | Hot-start polymerases remain inactive until a high-temperature step, dramatically reducing non-specific amplification and primer-dimer formation during reaction setup [10] [11]. |
| MgCl₂ or MgSO₄ Solution | The essential cofactor for DNA polymerase activity. Its concentration must be optimized for each primer-template system, as it directly influences enzyme processivity, primer annealing specificity, and fidelity [66] [11]. |
| PCR Additives (e.g., DMSO, Betaine) | These co-solvents help denature GC-rich templates and disrupt secondary structures that can cause polymerase stalling, thereby improving the amplification of difficult targets [66] [11]. |
| dNTP Mix | The building blocks for DNA synthesis. Using a high-quality, equimolar mix is vital for efficient amplification and to maintain low error rates during PCR [11]. |
| Nuclease-Free Water | The solvent for all reactions. It must be free of nucleases and PCR inhibitors to prevent degradation of templates and primers or inhibition of the polymerase [65]. |
| Standardized DNA Ladder | A critical reference for determining the size of amplified products on a gel, allowing for confirmation of the target amplicon and identification of non-specific products [61]. |
| Nucleic Acid Stain | Used to visualize DNA in gels. Sensitive fluorescent stains are preferred for detecting low-abundance products. Stains must be compatible with the gel type (e.g., denaturing for RNA) [61]. |
Technical Support & Troubleshooting Hub
Frequently Asked Questions (FAQs)
What is the fundamental role of MgCl₂ in a PCR reaction? MgCl₂ is an essential cofactor for DNA polymerase enzyme activity [1]. The magnesium ions (Mg²⁺) facilitate the catalytic function of the enzyme by binding to dNTPs and enabling the formation of phosphodiester bonds, which is crucial for the synthesis of new DNA strands [1]. Additionally, Mg²⁺ influences the melting temperature (Tm) of primers by neutralizing the negative charge on the DNA backbone, which promotes proper annealing between the primer and the template DNA [1].
What are the typical symptoms of suboptimal MgCl₂ concentration? The effects of incorrect MgCl₂ concentration are distinct [19] [67] [16]:
- Too High Concentration (e.g., >4.5 mM): Leads to non-specific binding of primers, resulting in multiple erroneous DNA products. This often appears as a smear or ladder of bands on an agarose gel [19] [16]. It also increases the risk of primer-dimer formation [19].
- Too Low Concentration (e.g., <1.5 mM): Prevents primers from binding effectively to the template, leading to greatly reduced yield or complete PCR failure with no visible amplification product [19] [67].
My gel shows a smear. Could MgCl₂ be the cause? Yes, a smeared appearance on an agarose gel is a classic indicator of non-specific amplification, which can be directly caused by an excessively high concentration of MgCl₂ [16]. Optimization of the MgCl₂ concentration is a primary troubleshooting step for this issue.
What is the standard starting range for MgCl₂ concentration? For most conventional PCR protocols, the ideal MgCl₂ concentration falls within a range of 1.5 mM to 4.5 mM, with 2.0 mM being a commonly used starting point [19] [67]. However, the optimal concentration must be determined empirically for each specific assay [1].
Troubleshooting Guide: Non-Specific Bands and Smearing
Problem: Agarose gel analysis of your PCR product reveals multiple non-specific bands or a diffuse smear, instead of a single, sharp band of the expected size.
Primary Suspect: Excessive MgCl₂ concentration is a leading cause.
Solution Pathway: A Stepwise Optimization Procedure
Step 1: Prepare a MgCl₂ Titration Master Mix Create a master mixture containing all PCR components except MgCl₂. Aliquot this master mix into multiple PCR tubes. Then, add a gradient of MgCl₂ volumes to each tube to test a range of final concentrations as outlined in the table below. This efficient one-experiment approach pinpoints the optimal condition [16].
Table: Experimental Setup for MgCl₂ Titration
| Tube | Target Final [MgCl₂] (mM) | Volume of 25 mM MgCl₂ Stock per 50 µL Reaction |
|---|---|---|
| 1 | 1.5 | 3.0 µL |
| 2 | 2.0 | 4.0 µL |
| 3 | 2.5 | 5.0 µL |
| 4 | 3.0 | 6.0 µL |
| 5 | 3.5 | 7.0 µL |
| 6 | 4.0 | 8.0 µL |
| 7 | 4.5 | 9.0 µL |
Step 2: Execute and Analyze the PCR
- Run the PCR using your standard thermocycling protocol.
- Analyze all reactions on the same agarose gel.
- Identify the MgCl₂ concentration that produces the strongest desired band with the least or no background smearing [16].
Step 3: Investigate Other Potential Causes If MgCl₂ titration does not resolve the issue, consider and troubleshoot these other common factors:
- Primer Design/Quality: Verify primer specificity and check for degradation [16].
- Template Quality & Quantity: Use high-quality, pure DNA and optimize the amount [16].
- Thermal Cycling Conditions: Optimize the annealing temperature [8].
- Enzyme Concentration: Ensure you are not using an excessive amount of DNA polymerase [16].
The following workflow diagram illustrates the logical process for troubleshooting smeared PCR products through MgCl₂ optimization:
Quantitative Data for MgCl₂ Optimization
The following table consolidates key quantitative relationships for MgCl₂ optimization, derived from recent research and meta-analyses.
Table: Evidence-Based Guidelines for MgCl₂ Optimization
| Parameter | Optimal Range / Value | Quantitative Effect & Context | Source |
|---|---|---|---|
| General Working Range | 1.5 - 4.5 mM | Standard starting range for most PCR protocols. | [19] [67] |
| Meta-Analysis Optimal Range | 1.5 - 3.0 mM | Identified as the statistically optimal range for balancing efficiency and specificity. | [15] |
| Effect on Melting Temp (Tₘ) | Increase of ~1.2 °C per 0.5 mM | Logarithmic relationship; every 0.5 mM increase within the optimal range raises DNA duplex Tₘ. | [15] |
| Template-Specific Needs | Higher concentrations for complex templates | Genomic DNA requires more MgCl₂ than simple plasmid/ cDNA templates. | [15] |
| dNTP Interaction | Critical co-optimization | Mg²⁺ binds dNTPs; therefore, the dNTP:primer interaction is a highly important variable. | [47] |
Advanced Protocols: Predictive Modeling for Standardization
Emerging research is moving beyond empirical "trial-and-error" optimization towards a standardized, predictive framework. The following protocol is based on a study that integrated thermodynamic modeling with machine learning to predict optimal MgCl₂ concentrations with high accuracy (R² = 0.9942) [47].
Protocol: Predictive MgCl₂ Concentration Modeling
Objective: To calculate a starting MgCl₂ concentration for a new assay using a predefined mathematical model, thereby reducing initial optimization time and reagents.
Theoretical Foundation: The model is based on a multivariate Taylor series expansion that incorporates thermodynamic principles (Gibbs free energy: ΔG = ΔH - TΔS) to account for molecular interactions between Mg²⁺ ions and DNA [47].
Materials & Reagents:
- Software: Python environment with scientific libraries (e.g., scikit-learn) [47].
- Input Variables: The following parameters must be known or calculated for your assay:
- Primer Melting Temperature (Tₘ)
- Primer GC Content (%GC)
- Amplicon Length (L) in base pairs
- dNTP Concentration (dNTP)
- Primer Concentration (Primers)
- Polymerase Concentration (Polymerase)
- Reaction pH
Predictive Equation: The study derived the following equation for predicting MgCl₂ concentration [47]: (MgCl₂) ≈ 1.5625 + (-0.0073 × Tₘ) + (-0.0629 × GC) + (0.0273 × L) + (0.0013 × dNTP) + (-0.0120 × Primers) + (0.0007 × Polymerase) + (0.0012 × log(L)) + (0.0016 × TₘGC) + (0.0639 × dNTPPrimers) + (0.0056 × pH_Polymerase)
Procedure:
- Data Collection: Compile all required input variables for your specific PCR assay.
- Calculation: Input the variables into the predictive equation.
- Empirical Validation: Use the calculated MgCl₂ value as the center point for a small-scale titration experiment (e.g., testing ±0.5 mM) to confirm prediction accuracy.
The workflow below visualizes this advanced, standardized optimization approach:
The Scientist's Toolkit: Essential Research Reagents
Table: Key Reagents for PCR and MgCl₂ Optimization
| Reagent / Solution | Critical Function in PCR |
|---|---|
| MgCl₂ Stock Solution (25 mM) | Standard stock concentration used for titration; provides the Mg²⁺ cofactor essential for polymerase activity and primer annealing [1] [16]. |
| Thermostable DNA Polymerase (e.g., Taq) | Enzyme that synthesizes new DNA strands; its activity is directly dependent on Mg²⁺ concentration [1] [8]. |
| dNTP Mix | The building blocks (dATP, dCTP, dGTP, dTTP) for new DNA synthesis. Mg²⁺ binds to dNTPs to form the active substrate for the polymerase, making dNTP concentration a key variable to co-optimize with MgCl₂ [47] [1]. |
| PCR Buffer (Mg²⁺-Free) | Provides the optimal chemical environment (pH, ionic strength) for the reaction. Using a Mg²⁺-free buffer is essential for performing a definitive MgCl₂ titration experiment [8]. |
| Universal Reporters / Hydrolysis Probes | For real-time PCR assays; probe efficiency can be significantly influenced by Mg²⁺ concentration and design factors like dimer stability, requiring integrated optimization [68]. |
Conclusion
Optimizing MgCl2 concentration is a cornerstone of robust PCR assay development, directly impacting the specificity, efficiency, and reliability of amplification. As detailed in this guide, moving from empirical troubleshooting to a science-driven approach—informed by an understanding of Mg2+ biochemistry, systematic titration, and even predictive modeling—empowers researchers to consistently eliminate nonspecific bands. For the fields of drug development and clinical diagnostics, where assay reproducibility is paramount, such rigorous optimization is not merely a technical step but a critical prerequisite. Future advancements will likely see greater integration of in-silico prediction tools with laboratory workflows, further streamlining the path to flawless PCR conditions and accelerating biomedical discovery.
References
- 1. What Is the Role of MgCl2 in PCR Amplification Reactions? [https://www.excedr.com/resources/what-is-the-role-of-mgcl2-in-pcr]
- 2. What is the mechanism of MgCl2 during PCR amplification? [https://www.aatbio.com/resources/faq-frequently-asked-questions/what-is-the-mechanism-of-mgcl2-during-pcr-amplification]
- 3. PCR Setup—Six Critical Components to Consider [https://www.thermofisher.com/us/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/pcr-education/pcr-reagents-enzymes/pcr-component-considerations.html]
- 4. Investigating concentration effects on reaction efficiency ... [https://www.sciencedirect.com/science/article/abs/pii/S0003269725001472]
- 5. What happens if I add too much MgCI2 in my PCR reaction? [https://www.aatbio.com/resources/faq-frequently-asked-questions/what-happens-if-i-add-too-much-mgci2-in-my-pcr-reaction]
- 6. Optimization of PCR conditions for amplification of GC-Rich ... [https://pubmed.ncbi.nlm.nih.gov/24218132/]
- 7. Help troubleshoot multiple Bands on gel (Conventional PCR) [https://biology.stackexchange.com/questions/115743/help-troubleshoot-multiple-bands-on-gel-conventional-pcr]
- 8. Polymerase Chain Reaction: Basic Protocol Plus ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4846334/]
- 9. Four tips for PCR amplification of GC-rich sequences [https://www.neb.com/en-us/nebinspired-blog/four-tips-for-pcr-amplification-of-gc-rich-sequences?srsltid=AfmBOoqZtJTD8_DlBS97QsQ7cc6IztymUlCag_wjU6awRAFiNQbY_TUo]
- 10. PCR Troubleshooting: Common Problems and Solutions [https://www.mybiosource.com/learn/pcr-troubleshooting-common-problems-and-solutions/]
- 11. PCR Troubleshooting Guide | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/pcr-education/pcr-reagents-enzymes/pcr-troubleshooting.html]
- 12. Four tips for PCR amplification of GC-rich sequences [https://www.neb.com/en-us/nebinspired-blog/four-tips-for-pcr-amplification-of-gc-rich-sequences?srsltid=AfmBOooH_LD9il3Z7sgqFh3vq9dUKJwPbG-KjCyUCkt1SVDq3S-ddZIN]
- 13. PCR Optimization: Factors Affecting Reaction Specificity ... [https://www.mybiosource.com/learn/pcr-optimization-factors-affecting-reaction-specificity-and-efficien/]
- 14. Setup—Six Critical Components to... | Thermo Fisher Scientific - RU PCR [https://www.thermofisher.com/ru/ru/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/pcr-education/pcr-reagents-enzymes/pcr-component-considerations.html]
- 15. Investigating concentration effects on reaction efficiency ... [https://pubmed.ncbi.nlm.nih.gov/40436087/]
- 16. Why do I get smeared PCR products? [https://www.qiagen.com/us/resources/faq/87]
- 17. Weak PCR Band Results or Smearing - What To Do [https://www.goldbio.com/blogs/articles/troubleshooting-pcr-part-three-solutions-for-weak-bands-and-smearing?srsltid=AfmBOortBQH9EUkpZLCZ6oZquGHxHhRQi72LskYeUXmFUejPeLi75OCp]
- 18. Clean Up Your Act! How To Clean Up PCR Contamination [https://bitesizebio.com/20773/clean-up-your-act-how-to-clean-up-pcr-contamination/]
- 19. How does magnesium concentration affect PCR? [https://www.aatbio.com/resources/faq-frequently-asked-questions/how-does-magnesium-concentration-affect-pcr]
- 20. Optimizing your PCR [https://www.takarabio.com/learning-centers/pcr/faq/optimization?srsltid=AfmBOop9zeddNg-m6SeM4ywD84_LdFG0vsrR6cA_oobZob1lsmAHrqmu]
- 21. Number of PCR Cycles and Magnesium Chloride ... [https://file.scirp.org/Html/6-2270410_49726.htm]
- 22. PCR Troubleshooting 101: How to Address Non-Specific ... [https://geneticeducation.co.in/pcr-troubleshooting-101-how-to-address-non-specific-amplification/]
- 23. Weak PCR Band Results or Smearing - What To Do [https://www.goldbio.com/blogs/articles/troubleshooting-pcr-part-three-solutions-for-weak-bands-and-smearing?srsltid=AfmBOorzyRuT4dmPfU48nhWA_RD8kUV4XyhTz1x6PP37VEdacAoLc2gg]
- 24. 5 Steps to a Better PCR: A Troubleshooting and ... [https://blog.abclonal.com/blog/5-steps-to-a-better-pcr]
- 25. 5 Essential Tips for Successful Gradient PCR [https://www.4esci.com/5-essential-tips-for-successful-gradient-pcr.html?srsltid=AfmBOorGI2bD0Z78dEqRK1ra042QzH_DWzuBMrlpPxZVhNutW3ZpZkJt]
- 26. Some Optimizing Strategies for PCR Amplification Success [https://www.creative-biogene.com/blog/some-optimizing-strategies-for-pcr-amplification-success]
- 27. Optimizing PCR: Proven Tips and Troubleshooting Tricks [https://www.the-scientist.com/optimizing-pcr-proven-tips-and-troubleshooting-tricks-71660]
- 28. Optimization of PCR Conditions for Amplification of GC‐Rich EGFR... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6807403/]
- 29. Impact of metal ions on PCR inhibition and RT- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7782418/]
- 30. Faster PCR Optimization [https://bitesizebio.com/31937/optimizing-your-pcr-faster/]
- 31. End-point PCR and PCR Primers Support - Troubleshooting [https://www.thermofisher.com/br/en/home/technical-resources/technical-reference-library/pcr-cdna-synthesis-support-center/end-point-pcr-primers-support/end-point-pcr-primers-support-troubleshooting.html]
- 32. Optimizing your PCR [https://www.takarabio.com/learning-centers/pcr/faq/optimization?srsltid=AfmBOoqloe3q5neBvBvRzHFlPBhepMKzaTIJx85S3H2ItEa241KoWn8o]
- 33. Getting No Bands in your PCR Result - Tips to Help [https://www.goldbio.com/blogs/articles/troubleshooting-pcr-part-two-what-to-do-when-you-are-not-getting-bands?srsltid=AfmBOorrAx_KV-N7TTSNxprc8gzMMQ1b6jkxYZd7QHS-yw81ewTxiSWf]
- 34. PCR Methods - Top Ten Strategies [https://www.thermofisher.com/us/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/pcr-education/pcr-reagents-enzymes/pcr-methods.html]
- 35. Touchdown PCR: Easy Intro Plus 5 Expert Tips [https://bitesizebio.com/2203/touchdown-pcr-a-primer-and-some-tips/]
- 36. How to optimize PCR conditions for GC-rich regions? [https://synapse.patsnap.com/article/how-to-optimize-pcr-conditions-for-gc-rich-regions]
- 37. PCR optimization - Molecular Biology [https://www.protocol-online.org/biology-forums/posts/8621.html]
- 38. Four tips for PCR amplification of GC-rich sequences [https://www.neb.com/en-us/nebinspired-blog/four-tips-for-pcr-amplification-of-gc-rich-sequences?srsltid=AfmBOorp8fCnrJljbt0Uh-DMsUk5lC94S8fK33P42wq1BoHqkBPOplQK]
- 39. What is the role of magnesium in PCR, and what ... [https://help.takarabio.com/36168/kb/article/110034/what-is-the-role-of-magnesium-in-pcr-and-what-is-the-optimal-concentration]
- 40. Effect of concentration of MgCl2 on random-amplified DNA ... [https://pubmed.ncbi.nlm.nih.gov/8024785/]
- 41. Four tips for PCR amplification of GC-rich sequences [https://www.neb.com/en-us/nebinspired-blog/four-tips-for-pcr-amplification-of-gc-rich-sequences?srsltid=AfmBOooe4n6Eh7LO91zkgdWocRick-O9LUatkOM0Clq2r57p2hRA8vIv]
- 42. PCR Optimization: Strategies To Minimize Primer Dimer [https://genemod.net/blog/pcr-optimization]
- 43. Four tips for PCR amplification of GC-rich sequences [https://www.neb.com/en-us/nebinspired-blog/four-tips-for-pcr-amplification-of-gc-rich-sequences?srsltid=AfmBOorL6p7X1M1Vj332WnJ_ZGK9xfewZlVcvlnO3-A-NH5QdfdDQCKK]
- 44. Optimizing your PCR [https://www.takarabio.com/learning-centers/pcr/faq/optimization?srsltid=AfmBOop0y7icuQD7ae8B3EtxFaGz6OhDfUzlhLtwwCJ-ackTMxyksypZ]
- 45. PCR Troubleshooting Guide [https://www.neb.com/en-us/tools-and-resources/troubleshooting-guides/pcr-troubleshooting-guide?srsltid=AfmBOoriycu9djgZeNTSFjEhKpbnSN6eTsYdiSNgibzV3LRi5PvrK-Yi]
- 46. Standard PCR Protocol [https://www.sigmaaldrich.com/US/en/technical-documents/protocol/genomics/pcr/standard-pcr?srsltid=AfmBOorKBsRgoAelzaJq58SS-aM7_pUolQnUAXHTYG7Z1uQrYeY303nz]
- 47. Mathematical Modeling and Thermodynamic Integration for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12620865/]
- 48. Troubleshooting Non-Specific Amplification [https://bento.bio/resources/bento-lab-advice/troubleshooting-non-specific-amplification-with-bento-lab/]
- 49. No Bands - Genotyping [https://www.jax.org/jax-mice-and-services/customer-support/technical-support/genotyping/troubleshooting/no-bands]
- 50. Magnesium Chloride (25 mM MgCl2) [https://bento.bio/resources/datasheets/magnesium-chloride-25-mm-mgcl2/]
- 51. Four tips for PCR amplification of GC-rich sequences [https://www.neb.com/en-us/nebinspired-blog/four-tips-for-pcr-amplification-of-gc-rich-sequences?srsltid=AfmBOoromTZsxhMlrmA73wXzN6IAH8aNYbkrJx3sFtPGFn1Ngk4hklzV]
- 52. Weak PCR Band Results or Smearing - What To Do [https://www.goldbio.com/blogs/articles/troubleshooting-pcr-part-three-solutions-for-weak-bands-and-smearing?srsltid=AfmBOoo0W9XXOCqEl75wk72_UGW7KfjqBhyxgNEs0-LlyllHn4DaxVqj]
- 53. PCR Basic Troubleshooting Guide [https://www.creative-biogene.com/blog/pcr-basic-troubleshooting-guide]
- 54. Investigating concentration effects on reaction efficiency ... [https://www.sciencedirect.com/science/article/pii/S0003269725001472]
- 55. Four tips for PCR amplification of GC-rich sequences [https://www.neb.com/en-us/nebinspired-blog/four-tips-for-pcr-amplification-of-gc-rich-sequences?srsltid=AfmBOornTDlH1GoxWvhd_UNxU9MjNP1w3X2wYPiR5SJK1sEFrCS1nIu3]
- 56. Measurement Errors in Polymerase Chain Reaction Are a ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0088031]
- 57. PCR Troubleshooting Guide [https://www.neb.com/en-us/tools-and-resources/troubleshooting-guides/pcr-troubleshooting-guide?srsltid=AfmBOooAN3ectFT3d9eteClmjV6b_0lUrh6AUZwTgkZ5r6A7L2jOSndc]
- 58. A Machine Learning Platform for Rapid PCR Assay Design ... [https://www.mdpi.com/2075-4418/15/12/1445]
- 59. What is the ideal concentration of MGCI2 in a PCR reaction? [https://www.aatbio.com/resources/faq-frequently-asked-questions/what-is-the-ideal-concentration-of-mgci2-in-a-pcr-reaction]
- 60. Standard PCR Protocol [https://www.sigmaaldrich.com/US/en/technical-documents/protocol/genomics/pcr/standard-pcr?srsltid=AfmBOop817N66IgF8dpWenMEO0xQQveJDOtDAAlYTs62MCBivROG5WGD]
- 61. Nucleic Acid Gel Electrophoresis Troubleshooting [https://www.thermofisher.com/us/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/na-electrophoresis-education/na-electrophoresis-troubleshooting.html]
- 62. Weak PCR Band Results or Smearing - What To Do [https://www.goldbio.com/blogs/articles/troubleshooting-pcr-part-three-solutions-for-weak-bands-and-smearing?srsltid=AfmBOooQNx7Ohx5KR52VNuWclzktZ-uCscnE5VE5HSbcjmvdSNoNo6O6]
- 63. Consensus guidelines for the validation of qRT-PCR ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8792405/]
- 64. Quantitative PCR validation for research scientists: Part 1 of 2 [https://virologyresearchservices.com/2023/01/13/quantitative-pcr-validation-part-1-of-2/]
- 65. Tips for successful PCR and Troubleshooting PCR-qPCR [https://info.abmgood.com/polymerase-chain-reaction-pcr-qpcr-troubleshooting]
- 66. Four tips for PCR amplification of GC-rich sequences [https://www.neb.com/en/nebinspired-blog/four-tips-for-pcr-amplification-of-gc-rich-sequences?srsltid=AfmBOool4fhtAE_SVFGeLrYlSk2SVPzPOIQRPbULWYGI2TcB_mQFXlV-]
- 67. What is the ideal concentration of MgCl2? [https://www.aatbio.com/resources/faq-frequently-asked-questions/what-is-the-ideal-concentration-of-mgcl2]
- 68. Real-time PCR probe optimization using design of ... [https://www.sciencedirect.com/science/article/pii/S2214753515300139]