NDR1 Nuclear vs. NDR2 Cytoplasmic Localization: Mechanisms, Functional Divergence in Signaling and Disease, and Research Applications
The homologous serine/threonine kinases NDR1 and NDR2, despite their high sequence similarity, exhibit a fundamental divergence in subcellular localization that dictates their non-overlapping functions in health and disease.
NDR1 Nuclear vs. NDR2 Cytoplasmic Localization: Mechanisms, Functional Divergence in Signaling and Disease, and Research Applications
Abstract
The homologous serine/threonine kinases NDR1 and NDR2, despite their high sequence similarity, exhibit a fundamental divergence in subcellular localization that dictates their non-overlapping functions in health and disease. NDR1 is predominantly nuclear, while NDR2 is primarily cytoplasmic and membrane-associated. This article provides a comprehensive analysis for researchers and drug development professionals, exploring the molecular mechanisms behind this differential localization and its profound functional consequences. We detail how this partitioning regulates distinct cellular processes—from innate immunity and antiviral response to synaptic plasticity, microglial activation, and cell cycle control. The content further covers advanced methodological approaches for studying these kinases, common troubleshooting pitfalls, and a direct comparative analysis of their roles in specific pathways like the Hippo signaling network. Understanding this NDR1/2 functional dichotomy is critical for developing targeted therapeutic strategies for cancer, inflammatory diseases, diabetic retinopathy, and neurodegenerative disorders.
Unraveling the NDR1/2 Dichotomy: Molecular Determinants of Nuclear and Cytoplasmic Partitioning
Nuclear Dbf2-related (NDR) kinases NDR1 (STK38) and NDR2 (STK38L) represent a conserved subclass of the AGC (protein kinase A/G/C) family of serine/threonine kinases, sharing approximately 87% amino acid sequence identity [1] [2]. These kinases are evolutionarily conserved from yeast to humans and belong to the NDR/LATS subfamily, which forms a crucial component of the Hippo signaling pathway [3]. While they perform overlapping functions in various cellular processes including cell cycle progression, apoptosis, and centrosome duplication, emerging evidence reveals critical functional specializations rooted in their distinct subcellular localization patterns [4] [5]. This comparative analysis delineates the core identity of NDR1 and NDR2 by examining their distribution, activation mechanisms, and specialized biological functions, providing researchers with structured experimental data and methodological guidance for investigating these kinases in physiological and pathological contexts.
Structural Similarities and Localization Differences
Despite their high degree of sequence similarity, NDR1 and NDR2 exhibit markedly different subcellular distributions that underpin their functional specialization. NDR1 is characterized primarily by its nuclear localization, while NDR2 displays a punctate cytoplasmic distribution [1] [6]. This fundamental difference in localization patterns was consistently observed across multiple cell types and experimental systems, suggesting distinct functional roles for these highly homologous kinases.
Table 1: Fundamental Characteristics of NDR1 and NDR2
| Feature | NDR1 (STK38) | NDR2 (STK38L) |
|---|---|---|
| Amino Acid Identity | ~87% shared identity | ~87% shared identity |
| Subcellular Localization | Predominantly nuclear [3] [1] | Punctate cytoplasmic distribution [4] [1] |
| Tissue Expression | Widely expressed [1] | Highest expression in thymus; widely expressed [1] |
| Peroxisomal Targeting | Lacks functional PTS1; diffuse distribution [4] | Contains C-terminal GKL motif; peroxisomal localization [4] [7] |
| PTS1 Receptor (Pex5p) Binding | No binding detected [4] | Direct binding demonstrated [4] |
The mechanistic basis for this differential localization was elucidated through research revealing that NDR2 contains a C-terminal peroxisome-targeting signal type 1 (PTS1)-like sequence, Gly-Lys-Leu (GKL), which is absent in NDR1 (which terminates in Ala-Lys) [4]. This GKL motif enables NDR2 to bind to the PTS1 receptor Pex5p, facilitating its recruitment to peroxisomes [4] [7]. Mutational studies confirmed this mechanism, as deletion of the C-terminal leucine in NDR2 (NDR2-ΔL) resulted in loss of punctate localization and diffuse cellular distribution similar to NDR1 [4].
Experimental Protocols for Studying Localization and Function
Subcellular Localization Assessment
Protocol 1: Immunofluorescence Microscopy for NDR1/NDR2 Localization
- Cell Culture: Plate human telomerase-immortalized retinal pigment epithelial (RPE1) or HeLa cells on glass coverslips in appropriate growth medium [4].
- Transfection: Transfect cells with plasmids encoding YFP- or GFP-tagged NDR1, NDR2, or NDR2-ΔL using Lipofectamine 2000 or similar transfection reagents [4].
- Fixation and Staining: At 24-48 hours post-transfection, fix cells with 4% paraformaldehyde, permeabilize with 0.1% Triton X-100, and incubate with primary antibodies against organelle markers (e.g., catalase for peroxisomes, EEA1 for early endosomes, GM130 for Golgi) [4].
- Imaging and Analysis: Capture images using confocal microscopy and analyze co-localization using image analysis software (e.g., ImageJ with JaCoP plugin) [4]. NDR2 should show significant co-localization with peroxisomal markers but not with markers of other organelles.
Protocol 2: Subcellular Fractionation and Immunoblotting
- Cell Lysis: Harvest NDR2-expressing HeLa cells and homogenize in isotonic buffer [4].
- Fractionation: Centrifuge post-nuclear supernatant at high speed to separate organellar (pellet) and cytosolic (supernatant) fractions [4].
- Density Gradient Centrifugation: Further separate the post-nuclear supernatant using iodixanol density gradient ultracentrifugation [4].
- Analysis: Collect fractions and analyze by SDS-PAGE and immunoblotting for NDR2 and organelle markers (e.g., Pex14p for peroxisomes) [4]. NDR2 should co-sediment with peroxisomal markers in specific density fractions.
Functional Interaction Studies
Protocol 3: Co-immunoprecipitation for NDR-MOB Interactions
- Cell Preparation: Culture Jurkat T-cells or HeLa cells and transfect with epitope-tagged NDR1, NDR2, and MOB constructs [1] [6].
- Cell Lysis: Lyse cells in non-denaturing lysis buffer (e.g., 1% NP-40, 150 mM NaCl, 50 mM Tris pH 8.0) with protease and phosphatase inhibitors [6].
- Immunoprecipitation: Incubate lysates with anti-epitope tag antibodies (e.g., anti-HA, anti-myc) followed by protein A/G beads [6].
- Analysis: Wash beads, elute bound proteins, and analyze by SDS-PAGE and immunoblotting for NDR and MOB proteins [6]. This protocol confirms physical interaction between NDR kinases and their regulatory MOB proteins.
Protocol 4: Kinase Activation Assay
- Reconstitution System: Express membrane-targeted versions of NDR and MOB proteins in COS-7, U2-OS, or HEK 293 cells [8].
- Stimulation: Treat cells with okadaic acid (1 μM, 60 minutes) to inhibit protein phosphatase 2A and enhance NDR phosphorylation [8].
- Phosphorylation Detection: Use phospho-specific antibodies against Ser281/Ser282 and Thr444/Thr442 of NDR1/NDR2 to assess activation status [8].
- Kinase Activity Measurement: Immunoprecipitate NDR kinases and perform in vitro kinase assays using specific substrate peptides [2] [9].
Activation Mechanisms and Signaling Pathways
NDR kinases require phosphorylation for full activation and are regulated by a conserved mechanism involving MOB proteins and upstream kinases. Both NDR1 and NDR2 undergo phosphorylation at two critical sites: a conserved threonine residue in the activation loop (Thr444 in NDR1, Thr442 in NDR2) and a serine residue for autophosphorylation (Ser281 in NDR1, Ser282 in NDR2) [8]. The association with MOB proteins (hMOB1A, hMOB1B, and hMOB2) dramatically stimulates NDR1 and NDR2 catalytic activity [1] [6].
Table 2: Activation Mechanisms and Regulatory Components
| Activation Parameter | NDR1 | NDR2 |
|---|---|---|
| Critical Phosphorylation Sites | Ser281, Thr444 [8] | Ser282, Thr442 [8] |
| Upstream Activators | MST kinases, MOB proteins [2] [9] | MST kinases, MOB proteins [2] |
| Subcellular Site of Activation | Nucleus, cytoplasm [8] | Primarily at plasma membrane [8] |
| Activation by Membrane Targeting | Constitutively active when membrane-targeted [8] | Constitutively active when membrane-targeted [8] |
| MOB Protein Binding | Binds hMOB1, hMOB2; dramatically stimulates activity [1] [9] [6] | Binds hMOB1, hMOB2; dramatically stimulates activity [1] [6] |
Research has demonstrated that membrane targeting of either NDR kinase results in constitutive activation due to phosphorylation at both regulatory sites, and this activation is further enhanced by co-expression of MOB proteins [8]. The activation of NDR kinases by membrane-bound MOBs occurs rapidly within minutes after MOB translocation to membranous structures [8].
Diagram 1: NDR Kinase Activation Pathway. NDR1/2 are activated through phosphorylation by upstream kinases (MST) and binding to MOB cofactors, which releases autoinhibition. Protein phosphatase 2A (PP2A) negatively regulates this pathway. Activated NDR1/2 phosphorylate downstream substrates to control diverse cellular functions.
Functional Specialization in Physiological Processes
The distinct subcellular localization of NDR1 and NDR2 translates to specialized physiological functions, despite their biochemical similarity.
NDR2 in Ciliogenesis and Peroxisomal Function
NDR2 plays a critical role in primary cilium formation, a function not shared with NDR1 [4] [7]. This specialized function depends on NDR2's peroxisomal localization mediated by its C-terminal GKL motif [4]. In ciliogenesis, NDR2 phosphorylates Rabin8, a GDP/GTP exchange factor for Rab8 GTPase, promoting local activation of Rab8 in the vicinity of the centrosome, which is essential for ciliary vesicle formation and axoneme growth [4] [2]. Functional studies demonstrated that expression of wild-type NDR2, but not the peroxisome-non-targeting mutant NDR2(ΔL), rescues the suppressive effect of NDR2 knockdown on ciliogenesis [4]. Furthermore, knockdown of peroxisome biogenesis factors (PEX1 or PEX3) partially suppresses ciliogenesis, confirming the importance of peroxisomal localization for NDR2's function in this process [4].
Differential Roles in Innate Immunity and Inflammation
NDR1 and NDR2 play distinct but complementary roles in regulating immune responses:
NDR1 functions as a negative regulator of TLR9-mediated immune response in macrophages by promoting ubiquitination and degradation of MEKK2, thereby inhibiting CpG-DNA-induced ERK1/2 activation and subsequent production of TNF-α and IL-6 [3]. NDR1 also acts as a transcriptional regulator of miR146a, dampening its transcription to promote STAT1 translation and enhance antiviral immune response [3].
NDR2 promotes RIG-I-mediated antiviral response by directly associating with RIG-I and TRIM25, facilitating complex formation and enhancing K63-linked polyubiquitination of RIG-I [3].
This functional specialization in immune regulation demonstrates how the differential localization of NDR1 and NDR2 enables them to participate in distinct signaling pathways, with NDR1 primarily modulating inflammatory cytokine production and NDR2 enhancing antiviral responses.
Neuronal Development and Function
Both NDR1 and NDR2 are expressed in the brain and contribute to neuronal development, but they may have distinct functions in this context [2] [5]. Research using chemical genetics identified AAK1 and Rabin8 as NDR1/2 substrates in the brain [2]. NDR1/2 kinases limit dendrite branching and length in cultured hippocampal neurons and in vivo, and are required for dendritic spine development and excitatory synaptic function [2]. Loss of NDR1/2 function leads to more immature spines and reduced frequency of miniature excitatory postsynaptic currents [2].
Table 3: Functional Specialization of NDR1 and NDR2 in Physiological Processes
| Biological Process | NDR1 Function | NDR2 Function |
|---|---|---|
| Ciliogenesis | No apparent role [4] | Critical role; phosphorylates Rabin8 to promote ciliary vesicle formation [4] |
| Centrosome Duplication | Involved [3] | Involved [3] |
| TLR9 Signaling | Negative regulator; targets MEKK2 for degradation [3] | Similar negative regulation based on siRNA studies [3] |
| Antiviral Immunity | Positive regulator; enhances STAT1 translation [3] | Positive regulator; enhances RIG-I ubiquitination [3] |
| Neuronal Development | Limits dendrite branching and length [2] | Limits dendrite branching and length [2] |
| Synaptic Function | Required for spine development [2] | Required for spine development [2] |
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagents for NDR1/2 Investigations
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Cell Lines | COS-7, U2-OS, HEK 293, HeLa, RPE1, Jurkat T-cells [8] [4] [1] | Model systems for localization, interaction, and functional studies |
| Expression Constructs | Epitope-tagged NDR1/2 (HA, myc, YFP, GFP), membrane-targeted versions, nucleus-targeted versions [8] [4] | Investigating localization, activation mechanisms, and functional consequences |
| Antibodies | Anti-NDR CT, Anti-NDR NT, phospho-specific antibodies (Ser281, Thr444) [8] | Detection, quantification, and localization of NDR kinases and activation status |
| Activation Reagents | Okadaic acid (PP2A inhibitor), 12-O-tetradecanoylphorbol 13-acetate (TPA) [8] | Experimental activation of NDR kinase pathways |
| Mutant Constructs | Kinase-dead (K118A, S281A/T444A), constitutively active, peroxisome-targeting deficient (NDR2-ΔL) [4] [2] | Functional dissection through gain-of-function and loss-of-function approaches |
| Interaction Partners | MOB1/2 expression constructs, Pex5p constructs [4] [1] | Studying regulatory mechanisms and pathway interactions |
| CCMQ | CCMQ (Constitution in Chinese Medicine Questionnaire) | The CCMQ is a 60-item tool for research on TCM body constitution types. This product is for Research Use Only (RUO). Not for personal use. |
| ML328 | ML328, MF:C22H21F3N6O3S, MW:506.5 g/mol | Chemical Reagent |
Pathological Implications and Research Directions
Dysregulation of NDR kinases has been implicated in various disease processes. NDR1 demonstrates tumor suppressor properties, with NDR1 knockout mice showing increased susceptibility to T-cell lymphoma [4] [5]. In contrast, NDR2 appears to function as an oncogene in certain cancers, particularly lung cancer, where it regulates processes including proliferation, apoptosis, migration, and invasion [10]. In the nervous system, NDR2 has been identified as the causal gene for canine early retinal degeneration, corresponding to the human ciliopathy Leber congenital amaurosis [4], highlighting its critical role in sensory neuronal maintenance.
The distinct subcellular localization patterns of NDR1 and NDR2—with NDR1 predominantly nuclear and NDR2 associated with peroxisomes—represent a fundamental aspect of their functional specialization. This differential targeting enables these highly similar kinases to participate in non-overlapping cellular processes while maintaining shared functions in other contexts. Future research should focus on further elucidating the complete interactomes of both kinases, developing more specific inhibitors, and exploring the therapeutic potential of targeting their distinct localization mechanisms in disease contexts, particularly cancer and neurological disorders.
Diagram 2: Functional Specialization and Pathological Implications. The distinct subcellular localization of NDR1 (nuclear) and NDR2 (peroxisomal) underlies their specialized cellular functions and associated disease implications when dysregulated.
The Nuclear Dbf2-Related (NDR) kinases NDR1 (STK38) and NDR2 (STK38L) are serine/threonine kinases belonging to the NDR/LATS subclass of AGC kinases, with crucial yet distinct roles in cellular processes including cell proliferation, apoptosis, morphogenesis, and immune regulation [4] [11]. Despite sharing 87% amino acid sequence identity and similar substrate specificities, these homologous kinases exhibit strikingly different subcellular localizations that dictate their non-overlapping functions in health and disease [4] [8].
NDR1 predominantly localizes to the nucleus, while NDR2 displays cytoplasmic distribution with specific organelle association [4] [8] [11]. This fundamental difference represents a fascinating molecular paradox: how do two highly similar proteins achieve such distinct localization patterns? The answer lies in divergent structural motifs—NDR1's functional Nuclear Localization Signal (NLS) versus NDR2's cytoplasmic anchors—that direct them to separate cellular compartments, thereby enabling specialized functional roles in cellular signaling networks.
Table 1: Fundamental Characteristics of NDR1 and NDR2 Kinases
| Characteristic | NDR1 (STK38) | NDR2 (STK38L) |
|---|---|---|
| Amino Acid Sequence Similarity | 87% identity with NDR2 | 87% identity with NDR1 |
| Primary Subcellular Localization | Nuclear | Cytoplasmic (peroxisomal) |
| Key Localization Motif | Functional NLS (residues 265-276) | C-terminal GKL peroxisomal targeting signal |
| Conservation | Conserved from yeast to humans | Conserved from yeast to humans |
| Kinase Class | NDR/LATS subclass of AGC kinases | NDR/LATS subclass of AGC kinases |
Molecular Determinants of Differential Localization
NDR1's Nuclear Localization Signal (NLS)
NDR1 contains a canonical nuclear localization signal (NLS) at residues 265-276, which facilitates its active import into the nuclear compartment [8]. Intriguingly, this NLS sequence is largely conserved in NDR2, with only a single conservative amino acid change, yet fails to function effectively in nuclear import [8]. This suggests that the NDR2 sequence contains structural features or post-translational modifications that override the potential NLS function.
Experimental evidence demonstrates that when NDR1 is artificially targeted to the plasma membrane using the myristoylation/palmitylation motif of the Lck tyrosine kinase, it becomes constitutively active due to phosphorylation on Ser281 and Thr444 [8]. This membrane-targeted activation provides important insights into how subcellular localization directly regulates NDR1 kinase activity.
NDR2's Cytoplasmic Anchoring Mechanisms
NDR2 exhibits punctate cytoplasmic localization rather than the diffuse distribution observed with NDR1 [4]. Through systematic colocalization studies with organelle markers, researchers have demonstrated that NDR2 specifically localizes to peroxisomes—single-membrane organelles involved in metabolic pathways including β-oxidation of fatty acids and detoxification of reactive oxygen species [4].
The molecular basis for this peroxisomal targeting lies in a C-terminal tripeptide sequence Gly-Lys-Leu (GKL) that functions as a peroxisome-targeting signal type 1 (PTS1) [4]. This critical discovery explains NDR2's cytoplasmic retention despite its sequence similarity to NDR1. Several lines of experimental evidence confirm this mechanism:
- PTS1 Receptor Binding: NDR2, but not NDR1, binds to the PTS1 receptor Pex5p, which mediates import of proteins into peroxisomes [4]
- Mutational Analysis: An NDR2 mutant lacking the C-terminal leucine (NDR2(ΔL)) exhibits diffuse cellular distribution instead of punctate peroxisomal localization [4]
- Functional Rescue: Wild-type NDR2, but not the peroxisome-non-targeting NDR2(ΔL) mutant, rescues the suppressive effect of NDR2 knockdown on ciliogenesis [4]
Table 2: Molecular Mechanisms Governing NDR1 and NDR2 Localization
| Localization Mechanism | NDR1 | NDR2 |
|---|---|---|
| Primary Targeting Signal | NLS (residues 265-276) | C-terminal GKL motif |
| Signal Type | Nuclear Localization Signal | Peroxisomal Targeting Signal 1 (PTS1) |
| Receptor/Binding Partner | Importin family | Pex5p (PTS1 receptor) |
| Localization Disruption | Mutation of NLS residues | Deletion of C-terminal leucine (ΔL mutant) |
| Secondary Localization | Cytoplasmic (upon activation) | Cell periphery and tips of microglial processes |
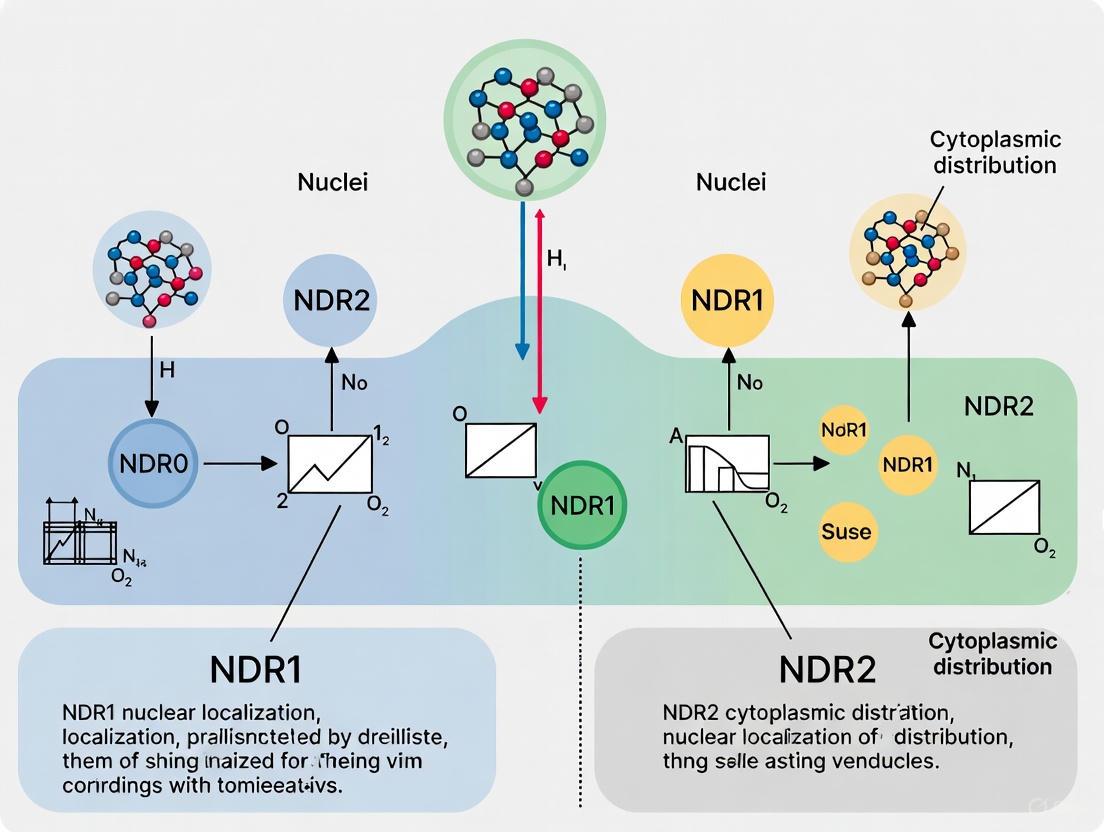
Diagram 1: Molecular determinants of NDR1 nuclear versus NDR2 cytoplasmic localization. NDR1 contains a functional NLS that directs nuclear import, while NDR2 possesses a C-terminal GKL motif that binds Pex5p for peroxisomal targeting.
Functional Consequences of Distinct Localization Patterns
NDR1's Nuclear Functions
NDR1's nuclear localization enables specific regulatory roles in gene expression and cell cycle control. As a nuclear kinase, NDR1 participates in phosphorylation and negative regulation of the transcriptional co-activator YAP1, thereby exerting tumor-suppressive functions [4]. Additionally, NDR1 plays a specialized role in regulating inflammatory responses through its nuclear actions, particularly in negatively regulating Toll-like receptor 9 (TLR9)-mediated immune responses in macrophages [11].
NDR1 deficiency leads to increased production of pro-inflammatory cytokines TNF-α and IL-6, indicating its critical role in preventing excessive inflammation induced by TLR signaling [11]. This nuclear-specific function highlights how NDR1's localization dictates its physiological role in immune regulation.
NDR2's Cytoplasmic Functions
NDR2's cytoplasmic and peroxisomal localization enables distinct functions in cellular morphogenesis and metabolic adaptation. A primary function of NDR2 is regulating primary cilium formation through phosphorylation of Rabin8, which promotes local activation of Rab8 GTPase in the vicinity of the centrosome [4]. This ciliogenesis function is specifically dependent on NDR2's peroxisomal localization, as demonstrated by the inability of peroxisome-non-targeting NDR2 mutants to rescue ciliogenesis defects [4].
In microglial cells, NDR2 plays a critical role in metabolic adaptation under high-glucose conditions, with NDR2 downregulation impairing mitochondrial respiration and reducing metabolic flexibility [12]. NDR2 also regulates cytoskeleton-dependent processes including phagocytosis and migration in microglial cells, consistent with its localization at the cell periphery and tips of cellular processes [12].
In disease contexts, NDR2's cytoplasmic functions become particularly significant. In non-small cell lung cancer (NSCLC), hypoxia-induced activation of NDR2 promotes brain metastasis by exacerbating HIF-1A, YAP, and C-Jun-dependent amoeboid migration [13]. NDR2 is more highly expressed in metastatic NSCLC than in localized NSCLC, and NDR2 silencing prevents xenograft formation and growth in lung cancer-derived brain metastasis models [13].
Table 3: Functional Specialization of NDR1 and NDR2 Based on Localization
| Functional Domain | NDR1 Nuclear Functions | NDR2 Cytoplasmic Functions |
|---|---|---|
| Cell Morphogenesis | Limited role | Central role in ciliogenesis via Rabin8 phosphorylation |
| Metabolic Regulation | Indirect through transcriptional control | Direct regulation of mitochondrial respiration and metabolic flexibility |
| Disease Processes | Tumor suppressor in colorectal cancer | Promotes metastasis in lung cancer |
| Immune Function | Negative regulation of TLR9 signaling | Regulation of microglial activation in diabetic retinopathy |
| Inflammatory Response | Competitively binds TRAF3 to promote IL-17 signaling | Facilitates breakdown of signaling molecules to inhibit IL-17 signaling |
Experimental Analysis of Localization Mechanisms
Key Methodologies for Localization Studies
Several experimental approaches have been crucial for deciphering the distinct localization patterns of NDR1 and NDR2:
Immunofluorescence and Confocal Microscopy: Researchers used immunocytochemistry with antibodies targeting specific regions of NDR kinases (N-terminus for NDR1/2 and C-terminus for NDR2) to demonstrate localization in human iPSC-derived microglial cultures, BV-2 immortalized microglial cells, and mouse primary retinal microglial cultures [12]. Colocalization studies with organelle-specific markers (catalase for peroxisomes, IBA1 for microglial processes) provided definitive evidence of subcellular distribution [12] [4].
Subcellular Fractionation and Biochemical Analysis: Biochemical fractionation of cellular components followed by Western blot analysis confirmed NDR2's association with peroxisomal fractions [4]. When post-nuclear supernatant fractions from YFP-NDR2-expressing HeLa cells were separated by iodixanol density gradient ultracentrifugation, NDR2 co-sedimented with the peroxisomal protein Pex14p [4].
Live-Cell Imaging and Mutational Analysis: The functional significance of localization motifs was tested through mutagenesis studies. Deletion of the C-terminal leucine in NDR2 (NDR2(ΔL)) resulted in diffuse cytoplasmic distribution instead of punctate peroxisomal localization [4]. Similarly, artificial targeting experiments using myristoylation/palmitylation motifs or nuclear localization signals demonstrated how localization affects kinase activity [8].
Diagram 2: Experimental workflow for determining NDR1 and NDR2 subcellular localization. Critical methodologies include tagged protein expression, cellular fractionation, immunofluorescence microscopy, and functional validation through mutational analysis.
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagents for Studying NDR1/NDR2 Localization and Function
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Antibodies | NDR1/2 antibody (E-2) #sc-271703; NDR2 antibody #STJ94368; IBA1 antibody | Detection and visualization of endogenous proteins and specific cellular markers |
| Cell Lines | BV-2 microglial cells; Human iPSC-derived microglia; HEK293; COS-7; HeLa | Model systems for localization studies and functional assays |
| Expression Constructs | YFP-NDR2; HA-NDR1; NDR2(ΔL) mutant; Membrane-targeted NDR | Manipulation of protein expression and localization for functional studies |
| Biochemical Assays | Subcellular fractionation kits; Co-immunoprecipitation reagents; Western blot systems | Analysis of protein localization, interactions, and activation status |
| Localization Markers | Catalase (peroxisomes); CFP-SKL (peroxisomes); Lamin A/C (nucleus); α-tubulin (cytoskeleton) | Reference standards for determining subcellular compartments |
| HUP30 | HUP30, CAS:312747-21-0, MF:C14H15N3O3S, MW:305.35 g/mol | Chemical Reagent |
| FQI1 | FQI1, CAS:599151-35-6, MF:C18H17NO4, MW:311.3 g/mol | Chemical Reagent |
Pathophysiological Implications and Therapeutic Perspectives
The distinct subcellular localization of NDR1 and NDR2 has significant implications for human disease and potential therapeutic interventions. In cancer biology, NDR1 often functions as a tumor suppressor, while NDR2 frequently exhibits oncogenic properties, particularly in lung cancer progression and metastasis [10] [13]. The hypoxia-induced activation of NDR2 in NSCLC and its role in promoting brain metastases highlights the clinical relevance of understanding these localization-specific functions [13].
In neurological contexts, NDR2 regulates microglial metabolic adaptation under high-glucose conditions relevant to diabetic retinopathy [12]. NDR2 downregulation impairs mitochondrial respiration, reduces phagocytic capacity, and elevates pro-inflammatory cytokines (IL-6, TNF, IL-17, IL-12p70), identifying NDR2 as a potential therapeutic target for neuroinflammatory processes [12].
The opposing roles of NDR1 and NDR2 in inflammatory signaling further demonstrate the functional significance of their distinct localizations. NDR1 promotes IL-17 signaling by competitively binding TRAF3, while NDR2 inhibits IL-17 signaling by facilitating the breakdown of signaling molecules [11]. This opposition suggests potential for targeted therapeutic interventions in autoimmune diseases by specifically modulating one kinase without affecting the other.
Future research directions should focus on developing small molecules that can specifically target one NDR kinase without affecting the other, potentially by exploiting their distinct subcellular localizations. Additionally, understanding the precise mechanisms that regulate the trafficking of each kinase between cellular compartments may reveal new opportunities for therapeutic intervention in cancer, inflammatory diseases, and metabolic disorders.
The differential subcellular localization of NDR1 and NDR2 represents a compelling example of how highly similar proteins evolve distinct functions through the acquisition of specific localization signals. NDR1's nuclear localization enables roles in transcriptional regulation and specific immune signaling pathways, while NDR2's cytoplasmic and peroxisomal targeting facilitates functions in ciliogenesis, metabolic adaptation, and cell migration. Understanding this "localization code" provides crucial insights for developing targeted therapeutic strategies that can selectively modulate the specific functions of each kinase in disease contexts. The continuing elucidation of NDR1 and NDR2 signaling networks will undoubtedly yield new opportunities for therapeutic intervention across a spectrum of human diseases.
The NDR (Nuclear Dbf2-related) kinase family, comprising NDR1 and NDR2, represents a crucial subclass of serine/threonine kinases within the conserved Hippo signaling pathway. These kinases, sharing approximately 87% amino acid sequence identity, play pivotal roles in regulating diverse cellular processes including cell proliferation, apoptosis, morphogenesis, and centrosome duplication [4] [14] [6]. Despite their high structural similarity, NDR1 and NDR2 exhibit distinct subcellular localization patterns and non-overlapping functions in specific biological contexts. NDR1 primarily localizes to the nucleus, while NDR2 displays a punctate cytoplasmic distribution [4] [6]. This functional divergence is particularly evident in primary cilium formation, where NDR2, but not NDR1, plays an essential role [4]. Understanding the structural determinants governing these differences requires a detailed comparative analysis of their N-terminal regulatory and C-terminal hydrophobic motifs, which serve as critical regulatory elements controlling kinase activity, subcellular targeting, and functional specificity.
Structural Organization of NDR Kinases
NDR kinases share a conserved domain architecture characteristic of the AGC group of serine/threonine kinases. Both isoforms contain a central kinase catalytic domain flanked by an N-terminal regulatory domain (NTR) and a C-terminal extension featuring a hydrophobic motif [14] [6]. The N-terminal regulatory region encompasses approximately 80 amino acids and includes five β-strands and one α-helix, forming a structurally conserved module that participates in autoinhibition and regulatory interactions [15] [16]. The C-terminal hydrophobic motif represents a crucial regulatory element that undergoes phosphorylation to modulate kinase activity [17].
Despite their high sequence conservation, key structural differences in both N-terminal and C-terminal regions dictate their distinct cellular functions and localization patterns. These variations affect interaction interfaces, phosphorylation susceptibilities, and subcellular targeting signals, ultimately contributing to the functional specialization of NDR1 and NDR2 in specific signaling contexts and physiological processes.
Table 1: Domain Architecture of NDR1 and NDR2 Kinases
| Domain/Region | NDR1 Features | NDR2 Features | Functional Consequences |
|---|---|---|---|
| N-terminal Regulatory Domain | Contains nuclear localization signals | Lacks strong nuclear localization signals | Dictates differential subcellular localization |
| Kinase Domain | Contains atypically long activation segment (∼30 aa) | Similar activation segment structure | Autoinhibition in NDR1; regulated by phosphorylation |
| C-terminal Hydrophobic Motif | Thr444 phosphorylation site | Thr442 phosphorylation site | Activation by upstream kinases (MST3) |
| C-terminal Tail | Ends with Ala-Lys | Ends with Gly-Lys-Leu (GKL) peroxisomal targeting signal | NDR2 localizes to peroxisomes; NDR1 does not |
Comparative Analysis of N-terminal Regulatory Motifs
The N-terminal regulatory domains of NDR kinases play critical roles in autoinhibition, protein-protein interactions, and subcellular targeting. Structural studies have revealed that the NDR1 kinase domain adopts an autoinhibited conformation stabilized by an atypically long activation segment that blocks substrate binding and stabilizes a non-productive position of helix αC [16]. This 30-amino acid insertion between kinase subdomains VII and VIII represents a key structural feature that maintains NDR1 in a basally inactive state, requiring conformational changes for full activation.
Structural determination of the human NDR1 kinase domain at 2.2 Å resolution has provided detailed insights into its autoinhibition mechanism. The extended activation segment forms extensive contacts with both the N-lobe and C-lobe of the kinase domain, preventing productive substrate binding and orienting helix αC in an inactive configuration [16]. Mutational analysis within this activation segment dramatically enhances NDR1 catalytic activity, confirming its autoinhibitory function. Specifically, residues within the β4-β5 turn and adjacent regions contribute to stabilization of the inactive state, with mutation of these elements resulting in constitutive kinase activation.
The N-terminal regions of NDR kinases also serve as critical interfaces for regulatory protein interactions. Both NDR1 and NDR2 bind to MOB proteins (MOB1A/B and MOB2), which dramatically stimulate their catalytic activities [6]. This interaction is mechanistically distinct from activation segment-mediated regulation, as MOB1 binding further potentiates the activity of NDR1 mutants with truncated autoinhibitory segments [16]. The N-terminal domain also facilitates interactions with upstream Hippo pathway components including MST1/2 kinases and the Furry (FRY) scaffold protein, creating a complex regulatory network that controls NDR kinase activity in response to diverse cellular signals.
Table 2: Key Functional Motifs in N-terminal Regulatory Domains
| Structural Element | NDR1 Characteristics | NDR2 Characteristics | Regulatory Consequences |
|---|---|---|---|
| Activation Segment | 30-amino acid insertion; autoinhibitory | Similar insertion; regulatory | Controls basal kinase activity; phosphoregulation |
| MOB Protein Binding Interface | High-affinity for MOB1A/B and MOB2 | High-affinity for MOB1A/B and MOB2 | Dramatic kinase activation upon binding |
| Furry Scaffold Binding Site | Direct interaction demonstrated | Presumed similar interaction | Facilitates upstream kinase recognition |
| N-lobe Regulatory Surfaces | Unique residue composition | Distinct residue patterns | Differential regulation by upstream inputs |
Comparative Analysis of C-terminal Hydrophobic Motifs
The C-terminal hydrophobic motifs of NDR kinases represent critical regulatory elements that undergo phosphorylation to control kinase activation. Both NDR1 and NDR2 contain conserved hydrophobic phosphorylation sites (Thr444 in NDR1 and Thr442 in NDR2) that are targeted by upstream Ste20-like kinases, particularly MST3 [17]. Phosphorylation of these residues results in a 10-fold stimulation of NDR kinase activity, with further enhancement mediated by MOB1A binding, leading to fully active kinases [17].
The mechanism of hydrophobic motif phosphorylation involves a multi-step activation process. For NDR2, MST3 selectively phosphorylates Thr442 in vitro, initiating a conformational change that facilitates subsequent autophosphorylation of the activation loop site Ser282 [17]. This ordered phosphorylation mechanism ensures precise control of NDR kinase activity in response to specific cellular signals. In vivo studies using kinase-dead MST3 mutants (MST3KR) demonstrate potent inhibition of Thr442 phosphorylation after okadaic acid stimulation, confirming MST3 as a bona fide upstream kinase for NDR2 hydrophobic motif phosphorylation [17].
Beyond their regulatory functions, the C-terminal regions of NDR kinases confer distinct subcellular localization properties. Notably, NDR2 contains a C-terminal peroxisome-targeting signal type 1 (PTS1)-like sequence (Gly-Lys-Leu), while NDR1 terminates with Ala-Lys [4]. This structural difference enables NDR2 to localize to peroxisomes through direct interaction with the PTS1 receptor Pex5p, while NDR1 lacks this targeting capability. Mutational studies confirm the functional importance of this motif, as deletion of the C-terminal Leu in NDR2 (NDR2(ΔL)) results in diffuse cytoplasmic distribution and impaired function in ciliogenesis [4].
Figure 1: NDR2 Activation and Peroxisomal Targeting Pathway. This diagram illustrates the multi-step activation process of NDR2 kinase through MST3-mediated phosphorylation and subsequent peroxisomal localization via Pex5p recognition of the C-terminal GKL motif.
Experimental Approaches for Structural-Functional Analysis
Subcellular Localization Studies
The distinct subcellular localization patterns of NDR1 and NDR2 have been characterized using fluorescence microscopy and subcellular fractionation techniques. Experimental protocols typically involve transfection of mammalian cells (such as RPE1 or HeLa cells) with plasmids encoding YFP- or CFP-tagged NDR constructs, followed by immunostaining with organelle-specific marker proteins [4]. For NDR2 peroxisomal localization, co-localization studies with established peroxisomal markers (catalase, CFP-SKL) provide definitive evidence. Quantitative analysis of fluorescence overlap using Pearson's correlation coefficient validates the specificity of observed localization patterns.
Subcellular fractionation protocols involve preparation of post-nuclear supernatant (PNS) fractions from transfected cells, followed by separation into organellar and cytosolic fractions through differential centrifugation [4]. Further purification using iodixanol density gradient ultracentrifugation enables precise determination of protein distribution across cellular compartments. Western blot analysis of fractionated samples using antibodies against NDR kinases and organelle-specific markers (e.g., Pex14p for peroxisomes) provides biochemical confirmation of microscopic observations.
Functional Mutagenesis and Chimera Studies
Structure-function relationships in NDR kinases have been elucidated through systematic mutagenesis and domain-swapping approaches. Site-directed mutagenesis of critical regulatory residues (e.g., Thr444/Thr442 in hydrophobic motifs, Ser281/Ser282 in activation loops) followed by in vitro kinase assays quantifies the contribution of specific residues to catalytic activity [17]. Experimental protocols typically involve expression of mutant kinases in mammalian cells (HEK293F, COS-7) or bacterial systems, immunopurification using epitope tags (HA, myc), and kinase activity measurements using specific substrates (e.g., histone H1) in the presence of [γ-32P]ATP.
Chimeric kinase constructs, generated by swapping specific domains or motifs between NDR1 and NDR2, enable precise mapping of functional determinants [15]. For example, grafting the β4-β5 turn region from Src to Csk has demonstrated the functional importance of this motif in kinase regulation [15]. Similarly, C-terminal truncation and point mutation experiments (e.g., NDR2(ΔL)) establish the role of specific sequences in subcellular targeting and functional specialization [4].
Interaction Studies and Phosphorylation Mapping
Protein-protein interactions involving NDR kinases have been characterized using co-immunoprecipitation and yeast two-hybrid approaches. Standard protocols involve co-transfection of epitope-tagged NDR and candidate binding partners (MOB proteins, Pex5p, RIG-I), followed by immunoprecipitation with tag-specific antibodies and Western blot analysis to detect associated proteins [4] [17] [6].
Phosphorylation status of specific regulatory sites has been monitored using phospho-specific antibodies (e.g., anti-P-Ser282, anti-P-Thr442) in combination with pharmacological inhibitors and RNA interference targeting upstream kinases [17]. For in vitro phosphorylation assays, recombinant kinases (MST3, NDR) are incubated with ATP, and phosphorylation reactions are analyzed by Western blotting or mass spectrometry to identify modification sites and quantify kinetics.
Table 3: Essential Research Reagents for NDR Kinase Studies
| Reagent Category | Specific Examples | Experimental Applications | Key Functions |
|---|---|---|---|
| Expression Constructs | pCMV5-HA-NDR1/2, pEGFP-NDR1/2, NDR2(ΔL) mutant | Localization, functional assays | Protein expression and mutagenesis studies |
| Antibodies | Anti-P-Ser282, Anti-P-Thr442, Anti-HA (12CA5), Anti-myc (9E10) | Western blot, immunoprecipitation | Detection of phosphorylation, protein expression |
| Cell Lines | HEK293F, COS-7, RPE1, HeLa | Kinase assays, localization studies | Cellular context for functional experiments |
| Biochemical Reagents | Okadaic acid, Microcystin, λ-phosphatase | Phosphatase inhibition/activation | Manipulating phosphorylation status |
| Interaction Partners | Recombinant MOB1A, MST3, Pex5p | In vitro binding and kinase assays | Studying regulatory mechanisms |
Functional Implications of Structural Differences
The structural variations in N-terminal and C-terminal motifs of NDR kinases have profound functional implications for their cellular roles. The peroxisomal localization of NDR2, mediated by its C-terminal GKL motif, directly links this kinase to primary cilium formation [4]. Functional studies demonstrate that wild-type NDR2, but not the peroxisome-targeting defective mutant NDR2(ΔL), rescues ciliogenesis defects in NDR2-knockdown cells. Furthermore, knockdown of peroxisome biogenesis factors (PEX1, PEX3) partially suppresses ciliogenesis, establishing a novel connection between peroxisomal signaling and cilium formation mediated by NDR2's unique C-terminal targeting motif [4].
The differential subcellular localization of NDR1 (nuclear) and NDR2 (cytoplasmic/peroxisomal) enables these kinases to participate in distinct signaling networks. NDR1 functions as a transcriptional regulator through binding to the intergenic region of miR146a, thereby modulating STAT1 translation and antiviral immune responses [14]. In contrast, NDR2 directly associates with cytoplasmic signaling complexes, such as the RIG-I/TRIM25 complex, enhancing K63-linked polyubiquitination of RIG-I and promoting antiviral type I interferon production [14]. These compartment-specific functions highlight how structural variations in regulatory motifs dictate functional specialization within the NDR kinase family.
Beyond their roles in innate immunity and ciliogenesis, the structural features of NDR kinases also influence their participation in cell cycle control, apoptosis, and Hippo pathway signaling. The autoinhibitory N-terminal extension in NDR1 provides an additional layer of regulation that may fine-tune its nuclear functions, while NDR2's peroxisomal targeting enables coordination between metabolic signaling and morphological changes. These functional specializations, rooted in structural differences, allow NDR1 and NDR2 to regulate diverse physiological processes while maintaining core kinase functions.
The comparative analysis of N-terminal regulatory and C-terminal hydrophobic motifs in NDR1 and NDR2 reveals how subtle structural variations generate functional diversity within a highly conserved kinase family. The extended N-terminal activation segment in NDR1 imposes autoinhibition that must be relieved through phosphorylation and MOB protein binding, while the C-terminal peroxisomal targeting motif in NDR2 directs this kinase to specific subcellular compartments where it regulates ciliogenesis. These structural differences, combined with variations in upstream activation mechanisms and binding partner interactions, enable NDR1 and NDR2 to perform non-redundant functions despite their high sequence similarity.
Understanding these structure-function relationships provides important insights for therapeutic targeting of NDR kinases in human diseases. The distinct subcellular localization patterns and activation mechanisms offer opportunities for isoform-specific modulation, potentially enabling selective intervention in pathological processes involving NDR signaling. Future structural studies, particularly focusing on full-length kinases in complex with their regulatory partners, will further elucidate the dynamic mechanisms governing NDR kinase function and regulation.
The nuclear Dbf2-related (NDR) kinases NDR1 (STK38) and NDR2 (STK38L) represent a crucial subfamily of AGC serine/threonine kinases that are highly conserved from yeast to humans [3]. These kinases function as key signaling nodes, integrating multiple regulatory inputs to control fundamental cellular processes including morphological changes, centrosome duplication, cell proliferation, apoptosis, and immune responses [3]. A defining characteristic of NDR kinase regulation involves a sophisticated mechanism dependent on phosphorylation at critical residues and interactions with MOB (Mps one binder) proteins. While NDR1 and NDR2 share approximately 87% sequence identity, they exhibit distinct subcellular localization patterns—NDR1 is primarily nuclear, whereas NDR2 is predominantly cytoplasmic [1]. This differential localization suggests non-redundant functions and potentially distinct activation mechanisms, forming a critical focus of ongoing research in the field. Understanding the precise molecular details of NDR kinase activation is essential not only for fundamental cell biology but also for therapeutic applications, given the roles of these kinases in cancer, neurodevelopment, and immune regulation [3] [2].
Molecular Machinery of NDR Kinase Activation
The Phosphorylation Code: Ser281 and Thr444
The activation mechanism of NDR1 hinges on phosphorylation at two conserved regulatory sites—Ser281 within the activation loop (T-loop) and Thr444 within the C-terminal hydrophobic motif (HM) [18] [19]. These phosphorylation events are essential for achieving full kinase activity and represent a convergence point for multiple upstream signals.
Ser281 undergoes autophosphorylation in a Ca²âº-dependent manner and serves as a key indicator of kinase activation [18]. Experimental evidence demonstrates that chelating intracellular Ca²⺠with BAPTA-AM suppresses both Ser281 phosphorylation and NDR1 activity, while treatments that increase cytoplasmic Ca²⺠concentrations enhance this autophosphorylation [18].
Thr444 is predominantly phosphorylated by an upstream kinase rather than through autophosphorylation [18]. Multiple kinases have been implicated in this regulatory step, including members of the MST kinase family (MST1-3) and PLK1, which phosphorylates NDR1 at mitotic entry to suppress its activity [20] [2]. Phosphorylation at both residues is indispensable for full NDR1 activation, as mutation of either site to alanine dramatically reduces kinase activity [18] [2].
Table 1: Critical Phosphorylation Sites in NDR1 Kinase
| Residue | Location | Phosphorylation Mechanism | Functional Consequence | Regulating Factors |
|---|---|---|---|---|
| Ser281 | Activation loop (T-loop) | Autophosphorylation | Induces kinase activation; essential for catalytic activity | Ca²âº, S100B binding, MOB proteins |
| Thr444 | Hydrophobic motif (HM) | Trans-phosphorylation by upstream kinases | Enables full kinase activation; regulates substrate binding | MST1/2, PLK1, MOB proteins |
| Thr74 | N-terminal region | Autophosphorylation | Facilitates S100B binding; minor regulatory role | Ca²⺠signaling |
MOB Proteins: Master Regulators of NDR Activity
MOB proteins function as critical cofactors that dictate NDR kinase activity through competitive binding and subcellular localization. The human genome encodes multiple MOB proteins, with MOB1 and MOB2 serving as the primary regulators of NDR kinases [1] [21].
MOB1 (including MOB1A and MOB1B isoforms) functions as a potent activator of NDR1/2. MOB1 binding to the N-terminal regulatory domain of NDR kinases stimulates kinase activity through multiple mechanisms: promoting autophosphorylation at Ser281, facilitating Thr444 phosphorylation by upstream kinases, and releasing autoinhibitory constraints within the catalytic domain [9] [19]. The association between MOB1 and NDR1 is particularly crucial for centrosome duplication, where it enables the formation of an MST1-MOB1-NDR1 signaling cascade [19].
MOB2 exhibits a more complex regulatory relationship with NDR kinases. While it binds to the same N-terminal domain as MOB1, it functions as a competitive inhibitor by preventing MOB1 binding and subsequent activation [21]. However, some contextual functions of MOB2 have been observed, particularly in neuronal development where it may contribute to NDR activation [2]. In hepatocellular carcinoma cells, MOB2 knockout promotes cell migration and invasion, while its overexpression has the opposite effect, suggesting tumor suppressor capabilities mediated through regulation of the Hippo pathway [21].
Table 2: MOB Protein Functions in NDR Kinase Regulation
| MOB Protein | Binding Partner | Effect on NDR Activity | Cellular Functions | Mechanistic Insights |
|---|---|---|---|---|
| MOB1A/B | NDR1/2, LATS1/2 | Strong activation | Centrosome duplication, mitotic progression, Hippo signaling | Releases autoinhibition, promotes Ser281 and Thr444 phosphorylation |
| MOB2 | NDR1/2 only | Competitive inhibition | Cell migration, cell cycle progression, DNA damage response | Prevents MOB1 binding; may fine-tune NDR activity in specific contexts |
Integrated Activation Mechanism
The current model of NDR1 activation involves an intricate sequence of molecular events: (1) initial binding of MOB1 to the N-terminal regulatory domain, which induces a conformational change that releases autoinhibition; (2) subsequent phosphorylation of Thr444 by upstream kinases such as MST1; and (3) autophosphorylation at Ser281, resulting in full kinase activation [19]. This multi-step mechanism ensures precise temporal and spatial control of NDR1 signaling, allowing integration of diverse cellular inputs.
Experimental Approaches and Key Methodologies
Assessing NDR Kinase Activity: Phosphospecific Antibodies and Kinase Assays
Research into NDR activation mechanisms relies heavily on well-established experimental protocols that enable precise monitoring of phosphorylation events and kinase activity.
Phosphospecific Antibody Detection: A fundamental methodology involves using phosphospecific antibodies that recognize NDR1 only when phosphorylated at specific residues. Antibodies targeting pSer281 and pThr444 have been extensively validated and provide a direct readout of activation status [8] [18] [20]. These tools have revealed that NDR1 phosphorylation at Thr444 is significantly reduced during mitosis, correlating with suppressed kinase activity [20]. Western blotting with these antibodies typically involves resolving proteins by SDS-PAGE (8-12% gels), transfer to PVDF membranes, blocking with 5% skim milk, and incubation with primary antibodies overnight followed by HRP-conjugated secondary antibodies and ECL detection [8].
In Vitro Kinase Assays: Direct measurement of NDR1 activity utilizes immunoprecipitated NDR1 incubated with [γ-³²P]-ATP and specific substrate peptides (e.g., KKRNRRLSVA) [19]. The transfer of radioactive phosphate to the substrate quantifies catalytic activity. This approach demonstrated that NDR1 isolated from interphase cells exhibits approximately 10-fold higher activity than NDR1 from mitotic cells [20]. Kinase assays using purified components have also established that MOB1 binding can dramatically stimulate NDR1 activity (up to 10-fold) in combination with phosphorylation events [1] [9].
Mutational Analysis: Dissecting Regulatory Mechanisms
Structure-function studies employing site-directed mutagenesis have been instrumental in deciphering NDR1 regulation:
- Kinase-dead mutants (K118A, S281A, T444A) abolish catalytic activity and function as dominant-negative variants [2] [19]
- Constitutively active mutants include the NDR1-PIF variant (containing the PRK2 hydrophobic motif) and NDR1EAIS with mutations in the autoinhibitory sequence [20] [19]
- Phospho-mimetic mutants (T444D/E) surprisingly do not recapitulate Thr444 phosphorylation, indicating that negative charge alone is insufficient to activate NDR1 [19]
- MOB-binding mutants with impaired MOB1 interaction help delineate MOB-dependent and MOB-independent functions [19]
Expression of these mutants in cellular models (e.g., Cos-7, U2-OS, HeLa cells) followed by phenotypic analysis has revealed that persistent NDR1 activation perturbs proper spindle orientation in mitosis, while kinase-dead NDR1 increases dendrite length and branching in neurons [20] [2].
Subcellular Localization and Imaging Studies
The distinct localization patterns of NDR1 (nuclear) and NDR2 (cytoplasmic) despite high sequence similarity represent a key area of investigation [1]. Experimental approaches include:
- Immunofluorescence microscopy using specific antibodies against endogenous NDR1/2 or epitope-tagged constructs [8] [20]
- Live-cell imaging with GFP-tagged NDR kinases to monitor dynamic localization during cell division [20]
- Inducible translocation systems where membrane-targeted MOB proteins rapidly recruit NDR to membranes, resulting in phosphorylation and activation within minutes [8]
These techniques have demonstrated that membrane targeting of NDR alone is sufficient to generate a constitutively active kinase due to spontaneous phosphorylation at both Ser281 and Thr444 [8].
Visualization of NDR1 Activation Pathway
The following diagram illustrates the integrated activation mechanism of NDR1 kinase, highlighting the key regulatory steps and molecular interactions:
Diagram Title: Integrated Activation Pathway of NDR1 Kinase
Key Regulatory Complexities
The activation pathway demonstrates several sophisticated regulatory features:
- Dual phosphorylation requirement: Both Ser281 and Thr444 phosphorylation must occur for full activation
- MOB competition: MOB1 and MOB2 compete for the same binding site, creating a toggle switch for activation
- Contextual upstream regulation: Different upstream kinases (MST1 vs PLK1) phosphorylate Thr444 depending on cellular context
- Calcium sensitivity: Ca²⺠signaling through S100B directly influences autophosphorylation capability
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Studying NDR Kinase Activation
| Reagent Category | Specific Examples | Research Applications | Key Characteristics |
|---|---|---|---|
| Phosphospecific Antibodies | Anti-pSer281-NDR1, Anti-pThr444-NDR1 | Monitoring activation status; Western blot, immunofluorescence | Validate kinase activation; assess regulation in different cellular states |
| Activation State Mutants | NDR1-K118A (kinase dead), NDR1-AA (S281A/T444A), NDR1-PIF (constitutively active) | Functional studies; pathway dissection | Determine necessity and sufficiency of NDR1 in cellular processes |
| MOB Expression Constructs | MOB1A/B (activator), MOB2 (inhibitor), membrane-targeted MOB variants | Mechanistic studies; pathway manipulation | Define MOB-specific functions; manipulate subcellular localization |
| Chemical Inhibitors/Activators | Okadaic acid (PP2A inhibitor), Thapsigargin (Ca²⺠mobilizer), BAPTA-AM (Ca²⺠chelator) | Probing regulatory mechanisms | Dissect phosphorylation dynamics; calcium dependence |
| Substrate Detection Systems | NDR substrate peptide (KKRNRRLSVA), AAK1, Rabin8 substrates | Kinase activity assays; substrate identification | Quantify enzymatic activity; identify downstream targets |
| Localization Tools | Centrosome-targeted NDR (AKAP-NDR1), NLS/NES tags, GFP-tagged NDR1/2 | Subcellular targeting studies | Investigate compartment-specific functions; live imaging |
| spb | `SPB Chemical Reagent|For Research Use Only` | SPB chemical reagent for research applications. This product is For Research Use Only (RUO). Not for human or veterinary use. | Bench Chemicals |
| UKI-1 | UKI-1|uPA System Inhibitor|CAS 220355-63-5 | UKI-1 is a novel, synthetic inhibitor of the urokinase plasminogen activator (uPA) system, investigated for solid tumor research. For Research Use Only. Not for human use. | Bench Chemicals |
Functional Consequences and Research Applications
The regulated activation of NDR kinases through MOB proteins and phosphorylation has profound implications for diverse biological processes:
Cell Division and Centrosome Duplication: Proper NDR1 activity control is essential for accurate mitotic progression. During mitosis, PLK1 phosphorylates NDR1 at three threonine residues (T7, T183, T407), which suppresses NDR1 activity by interfering with MOB1 binding [20]. Constitutively active NDR1 (NDR1EAIS) causes aberrant spindle rotation and orientation defects, highlighting the importance of temporal regulation [20].
Neuronal Development: NDR1/2 kinases limit dendrite length and proximal branching in mammalian pyramidal neurons [2]. Kinase-dead NDR1/2 mutants increase dendrite complexity, while constitutively active forms have the opposite effect. NDR1/2 also promotes dendritic spine maturation and excitatory synaptic function, with identified substrates including AAK1 (regulating dendrite growth) and Rabin8 (controlling spine development) [2].
Infection and Immunity: NDR1 plays complex roles in immune regulation, acting as a negative regulator of TLR9-mediated inflammation by promoting degradation of MEKK2, while positively regulating RIG-I-mediated antiviral response through enhancement of RIG-I/TRIM25 complex formation [3].
The experimental frameworks and reagents described in this guide provide researchers with comprehensive tools to further investigate the context-dependent functions of NDR kinases and their potential as therapeutic targets in cancer, neurological disorders, and inflammatory diseases.
The NDR (Nuclear Dbf2-related) kinase family represents a highly conserved subgroup of serine/threonine AGC kinases that function as crucial regulators of cell proliferation, apoptosis, morphogenesis, and cell polarity. These kinases are remarkably conserved throughout the eukaryotic domain, with orthologs identified from yeast to humans [22] [4] [23]. The founding member of this family, Dbf2, was first characterized in Saccharomyces cerevisiae, where it regulates mitotic exit and morphogenesis [6] [8]. In mammals, this family expanded to include four members: NDR1 (STK38), NDR2 (STK38L), LATS1, and LATS2, which form the core of the NDR/LATS kinase subfamily [24] [23]. The evolutionary conservation of these kinases extends beyond sequence homology to encompass their regulatory mechanisms and fundamental biological functions, making them a subject of intense research interest particularly in the context of differential subcellular localization and function between NDR1 (nuclear) and NDR2 (cytoplasmic).
Evolutionary Conservation Across Species
The NDR kinase family exhibits remarkable evolutionary conservation from lower eukaryotes to complex multicellular organisms. This conservation is evident not only in the amino acid sequences but also in their structural domains and activation mechanisms.
Table 1: NDR Kinase Orthologs Across Species
| Organism | NDR1/2 Ortholog | LATS1/2 Ortholog | Key Functions |
|---|---|---|---|
| S. cerevisiae (Budding yeast) | Cbk1p, Dbf2, Dbf20 | - | Mitotic exit, cell morphogenesis, cell integrity [23] |
| S. pombe (Fission yeast) | orb6, sid2 | - | Cell polarity, cytokinesis [23] |
| C. elegans (Nematode) | SAX-1 | WARTS/wts-1 | Neurite outgrowth, epidermal morphogenesis [4] [23] |
| D. melanogaster (Fruit fly) | Tricornered (Trc) | Warts (Wts) | Dendritic tiling, epidermal morphogenesis [4] [23] |
| H. sapiens (Human) | NDR1 (STK38), NDR2 (STK38L) | LATS1, LATS2 | Centrosome duplication, ciliogenesis, apoptosis, Hippo signaling [4] [24] [23] |
The sequence identity between human NDR1 and NDR2 is approximately 87%, highlighting their close relationship [4] [24]. Despite this high similarity, they exhibit distinct subcellular localizations—NDR1 is predominantly nuclear while NDR2 displays cytoplasmic distribution [4] [6]. This differential localization represents a key functional divergence that underscores their non-redundant biological roles.
Structural Conservation and Activation Mechanisms
NDR kinases share a conserved domain architecture consisting of an N-terminal regulatory domain (NTR) and a C-terminal kinase domain. A distinctive feature of all NDR kinases is an insert between catalytic subdomains VII and VIII characterized by high basic amino acid content [22]. Structural studies have revealed that this insert possesses autoinhibitory function, and binding of MOB proteins to the N-terminal domain induces conformational changes that release this autoinhibition [22].
The activation mechanism of NDR kinases is highly conserved and requires multiple phosphorylation events and regulatory interactions:
Phosphorylation Events
- Activation loop phosphorylation: Thr444 in NDR1 (Thr442 in NDR2) is phosphorylated by an upstream kinase [8]
- N-terminal phosphorylation: Ser281 in NDR1 (Ser282 in NDR2) involves autophosphorylation [8]
- Both phosphorylation events are essential for full kinase activation [8]
MOB Protein Interaction
MOB (Mps one binder) proteins serve as critical coactivators of NDR kinases. The interaction between NDR and MOB proteins is evolutionarily conserved from yeast to humans [22] [6]. Human MOB1 (hMOB1) directly binds to the N-terminal regulatory domain of NDR kinases, dramatically stimulating their catalytic activity [22] [6]. This interaction is so critical that membrane-targeted hMOBs can robustly promote NDR activation at the plasma membrane, demonstrating the functional significance of this conserved mechanism [8].
Diagram Title: Conserved NDR Kinase Activation Mechanism
Functional Divergence: NDR1 Nuclear Localization vs. NDR2 Cytoplasmic Distribution
Despite their high sequence similarity, NDR1 and NDR2 have evolved distinct subcellular localizations and functions, a key aspect of their functional divergence in mammalian systems.
NDR1 Nuclear Functions
NDR1 is predominantly localized in the nucleus and participates in critical nuclear processes:
- Cell cycle regulation: Controls centrosome duplication and mitotic chromosome alignment [4] [24]
- Transcriptional regulation: Phosphorylates and regulates transcriptional co-activators including YAP/TAZ in the Hippo pathway [4] [24] [23]
- Tumor suppression: Acts as a tumor suppressor in various cancers including prostate cancer and T-cell lymphoma [24] [25]
- Apoptosis regulation: Promotes apoptosis through phosphorylation of pro-apoptotic substrates [24] [25]
NDR2 Cytoplasmic Functions
NDR2 exhibits punctate cytoplasmic distribution and participates in distinct cellular processes:
- Ciliogenesis: Regulates primary cilium formation through phosphorylation of Rabin8, promoting local activation of Rab8 [4]
- Peroxisomal targeting: Localizes to peroxisomes using a C-terminal Gly-Lys-Leu (GKL) sequence that functions as a peroxisomal targeting signal (PTS1) [4]
- Vesicular trafficking: Controls trafficking of vesicles through regulation of Rab GTPase activity [4] [10]
- Metabolic adaptation: Regulates microglial metabolic adaptation under high-glucose conditions [12]
Table 2: Comparative Analysis of NDR1 and NDR2 Properties
| Property | NDR1 | NDR2 |
|---|---|---|
| Sequence Identity | Reference | 87% identical to NDR1 [4] [24] |
| Subcellular Localization | Diffuse nuclear and cytoplasmic [4] [6] | Punctate cytoplasmic; peroxisomal [4] |
| Tissue Distribution | Widely expressed [6] | Highest expression in thymus [6] |
| C-terminal Targeting Signal | Ala-Lys [4] | Gly-Lys-Leu (PTS1-like) [4] |
| Role in Ciliogenesis | Not essential [4] | Critical regulator [4] |
| Cancer Association | Tumor suppressor in prostate cancer, lymphoma [24] [25] | Oncogenic in lung cancer [10] |
| Immune Function | Regulates inflammation and immunity [25] | Modulates microglial inflammatory response [12] |
The mechanistic basis for their differential localization was elucidated by Hori et al. (2017), who discovered that NDR2 contains a C-terminal Gly-Lys-Leu (GKL) sequence that functions as a peroxisomal targeting signal (PTS1), while NDR1 terminates in Ala-Lys and does not localize to peroxisomes [4]. This critical difference explains the distinct subcellular distributions and functional specializations of these kinases.
Experimental Approaches and Key Methodologies
Research on NDR kinases employs sophisticated experimental approaches to elucidate their conservation, activation mechanisms, and functional differences.
Key Experimental Protocols
Subcellular Localization Studies
Protocol:
- Transfect cells with plasmids encoding N-terminally tagged YFP-NDR2 or YFP-NDR1 [4]
- Immunostain with antibodies against organelle markers (catalase for peroxisomes, EEA1 for early endosomes, GM130 for Golgi) [4]
- Analyze co-localization using fluorescence microscopy [4]
- Perform subcellular fractionation by iodixanol density gradient ultracentrifugation [4]
- Confirm peroxisomal localization through Pex5p binding assays [4]
Key Finding: NDR2, but not NDR1, co-localizes with peroxisomal markers and binds to the PTS1 receptor Pex5p [4].
Kinase Activation Assays
Protocol:
- Express and purify GST-fused NDR1 from E. coli BL21 using glutathione-agarose affinity chromatography [25]
- Incubate purified NDR1 with substrate peptide (KKRNRRLSVA), ATP, and reaction buffer [25]
- Treat with potential activators (e.g., MOB proteins or small-molecule agonists) [22] [25]
- Measure kinase activity using luminescent kinase assay kits [25]
- Assess phosphorylation status using phospho-specific antibodies [8]
Key Finding: MOB binding dramatically stimulates NDR kinase activity, and membrane targeting results in constitutive NDR activation [22] [8].
Functional Rescue Experiments
Protocol:
- Knock down endogenous NDR2 using siRNA or CRISPR-Cas9 [4] [12]
- Transfect with wild-type NDR2 or mutant NDR2(ΔL) lacking the C-terminal leucine [4]
- Assess functional recovery in ciliogenesis assays [4]
- Evaluate peroxisomal localization by immunofluorescence [4]
Key Finding: Wild-type NDR2, but not NDR2(ΔL), rescues ciliogenesis defects caused by NDR2 knockdown, establishing the functional significance of peroxisomal localization [4].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for NDR Kinase Studies
| Reagent/Tool | Function/Application | Example/Source |
|---|---|---|
| hMOB1A/B protein | NDR kinase co-activator for in vitro activation assays [22] | Recombinantly expressed [22] [8] |
| Phospho-specific antibodies | Detection of activated NDR (pThr444/442, pSer281/282) [8] | Custom-produced against phosphopeptides [8] |
| PTS1 receptor (Pex5p) | Investigation of NDR2 peroxisomal targeting mechanism [4] | Co-immunoprecipitation assays [4] |
| Okadaic acid (OA) | PP2A inhibitor used to activate NDR kinases by preventing dephosphorylation [8] | Commercial sources (Alexis Corp.) [8] |
| CRISPR-Cas9 plasmids | Generation of NDR knockout cell lines for functional studies [12] | sgRNA against exon 7 of Ndr2 gene [12] |
| Small-molecule agonists | Pharmacological activation of NDR kinases for therapeutic research [25] | aNDR1 compound [25] |
| 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid | 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid, CAS:7365-45-9, MF:C8H18N2O4S, MW:238.31 g/mol | Chemical Reagent |
| SN003 | SN003, MF:C19H25N5O2, MW:355.4 g/mol | Chemical Reagent |
Therapeutic Implications and Research Applications
The evolutionary conservation of NDR kinases extends to their relevance in human disease pathways, making them attractive therapeutic targets:
Cancer Therapeutics: NDR1 functions as a tumor suppressor in prostate cancer, with decreased expression correlating with poorer prognosis [24] [25]. Small-molecule NDR1 agonists like aNDR1 show promising antitumor activity both in vitro and in vivo [25].
Ciliopathies: NDR2's essential role in ciliogenesis implicates it in ciliopathies such as Leber congenital amaurosis, a form of early retinal degeneration [4].
Metabolic and Inflammatory Disorders: NDR2 regulates microglial metabolic adaptation under high-glucose conditions, suggesting potential applications in diabetic retinopathy [12].
Aging and Neurodegeneration: NDR kinases have been linked to various aging hallmarks including cellular senescence, chronic inflammation, and autophagy, positioning them as potential regulators of aging processes [23].
The evolutionary journey from yeast Dbf2 to mammalian NDR kinases represents a compelling narrative of conservation and divergence. While the core structure and activation mechanisms have been remarkably preserved across eukaryotic evolution, the mammalian NDR kinases have diversified into specialized functions exemplified by the distinct nuclear and cytoplasmic roles of NDR1 and NDR2. The differential subcellular localization of these kinases—governed by discrete targeting signals—underpins their functional specialization in regulating diverse cellular processes from centrosome function to ciliogenesis. Continued research on these evolutionarily conserved kinases promises not only to advance our fundamental understanding of cellular regulation but also to unlock novel therapeutic approaches for cancer, ciliopathies, metabolic disorders, and age-related diseases.
Advanced Techniques for Isolating and Probing NDR1- and NDR2-Specific Functions
The highly homologous kinases NDR1 and NDR2, despite significant sequence similarity, exhibit distinct subcellular localizations and biological functions that necessitate rigorous detection strategies. NDR1 contains a functional nuclear localization signal (NLS) and is found predominantly in the nucleus, while NDR2 is primarily cytoplasmic, a differential distribution that underpins their non-redundant roles in cellular processes [8]. This subcellular partitioning is functionally significant, with NDR1 implicated in cell cycle regulation at the G1/S transition and NDR2 playing crucial roles in antiviral immune responses, vesicular trafficking, and autophagy [26] [27] [10]. Accurate differentiation between these kinases through validated antibodies and carefully designed localization reporters is therefore paramount for advancing our understanding of their distinct functions in both physiological and pathological contexts, including cancer and neurodegenerative diseases [10] [28].
The challenge of specific detection is compounded by the high amino acid identity (>80%) between NDR1 and NDR2, which can lead to antibody cross-reactivity and misinterpretation of experimental results [10]. Furthermore, the expanding roles of NDR kinases in fundamental cellular processes and disease pathways underscores the critical need for reliable detection methodologies. This guide provides a comprehensive comparison of current strategies for the specific detection of NDR1 and NDR2, offering experimental protocols and analytical frameworks to empower researchers in generating reproducible and biologically relevant data.
Antibody Validation Strategies for Differentiating NDR1 and NDR2
The Five Pillars of Antibody Validation
Antibody validation is essential for ensuring specific, selective, and reproducible results in NDR kinase research. The following validation pillars provide a comprehensive framework for confirming antibody specificity in different experimental contexts [29]:
Genetic Knockout/Knockdown: This method involves using cells or organisms in which the gene encoding the target protein (NDR1 or NDR2) has been completely or partially inactivated. The specificity of an antibody is confirmed by a significant reduction or absence of signal in the knockout/knockdown samples compared to controls. For NDR kinases, this approach is particularly valuable due to their high similarity, requiring demonstration that an anti-NDR1 antibody does not recognize NDR2 in NDR1-knockout cells, and vice versa [29].
Orthogonal Validation: This strategy employs a non-antibody-based method to measure the same target protein. For NDR localization studies, this could involve comparing immunofluorescence results with genetically encoded fluorescent protein tags or mRNA detection techniques. Consistent results between the antibody-based method and the orthogonal approach provide strong validation evidence [29].
Use of Comparable Antibodies: Multiple antibodies raised against different epitopes of the same target protein should yield similar staining patterns. For NDR1 and NDR2, this would involve using antibodies targeting different regions of each kinase and confirming consistent subcellular localization patterns—nuclear for NDR1 and cytoplasmic for NDR2 [29] [8].
Immunoprecipitation Mass Spectrometry (IP/MS): An antibody is used to immunoprecipitate the target protein from a complex mixture, followed by identification of the pulled-down proteins via mass spectrometry. This method provides direct evidence for antibody specificity and can reveal potential off-target interactions, crucial for distinguishing between highly similar proteins like NDR1 and NDR2 [29].
Recombinant Protein Expression: Expressing the recombinant NDR1 or NDR2 protein in a heterologous system provides a positive control for antibody validation. The presence of a single band at the expected molecular weight in Western blot analysis confirms the antibody's specificity for its intended target [29].
Common Pitfalls in Antibody-Based Detection
Approximately 70% of researchers have struggled to reproduce experiments conducted by other scientists, often due to issues with antibodies [29]. Several critical pitfalls must be addressed when working with antibodies for NDR detection:
Nonspecific Antibodies: Antibodies may recognize unrelated epitopes or proteins with similar structures. This is particularly problematic for NDR1 and NDR2 due to their high sequence conservation. A study demonstrated that 35% of monoclonal antibody preparations analyzed had staining patterns unrelated to their intended antigenic specificity [30].
Non-reproducible Antibodies: Significant lot-to-lot variations can occur with commercial antibodies. One study highlighted two different lots of the same monoclonal antibody that showed completely different staining patterns—one nuclear and one membranous/cytoplasmic—with very poor correlation (R² = 0.038) [30].
Epitope Accessibility: The recognition of an antibody can be affected by protein conformation, post-translational modifications, and fixation methods. Epitopes accessible in denatured proteins for Western blot may be inaccessible in native proteins for immunohistochemistry, and vice versa [30].
Cellular Compartment Mislocalization: Staining patterns that contradict established biological knowledge indicate potential nonspecificity. For instance, cytoplasmic staining for a known nuclear transcription factor, or vice versa, should raise concerns about antibody validity [30].
Table 1: Troubleshooting Antibody Specificity for NDR Kinases
| Problem | Potential Cause | Solution |
|---|---|---|
| Unexpected bands in Western blot | Cross-reactivity with other proteins or modified forms | Validate by knockout/knockdown; optimize blocking conditions |
| Aberrant subcellular localization | Recognition of unrelated epitopes; fixation artifacts | Compare with orthogonal methods; optimize fixation protocol |
| High background noise | Non-specific antibody binding | Titrate antibody concentration; improve antigen retrieval |
| Inconsistent staining between lots | Variations in antibody production | Request validation data from vendor; test new lots extensively |
| Discrepancy between techniques | Differential epitope accessibility | Use multiple validation pillars; confirm antibody suitability for specific applications |
Experimental Protocol: Knockout Validation for NDR Antibodies
This protocol provides a robust method for validating NDR antibody specificity using genetic knockout cells:
Cell Culture: Maintain wild-type (WT) and NDR1 or NDR2 knockout (KO) cell lines (e.g., HEK293, HeLa, or U2OS) in appropriate medium supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 incubator [26] [8].
Sample Preparation:
- For Western blotting: Lyse cells in RIPA buffer containing protease and phosphatase inhibitors. Quantify protein concentration and resolve 20-30 μg by SDS-PAGE [8] [30].
- For immunofluorescence: Culture cells on glass coverslips, fix with 4% paraformaldehyde for 15 minutes, and permeabilize with 0.1% Triton X-100 in PBS for 10 minutes [8].
Immunodetection:
- For Western blotting: Transfer proteins to PVDF membranes, block with 5% non-fat milk in TBST, and incubate with primary antibodies against NDR1 or NDR2 (1:1000 dilution) overnight at 4°C. After washing, incubate with HRP-conjugated secondary antibodies (1:5000) for 1 hour at room temperature and detect using ECL reagent [8].
- For immunofluorescence: After blocking with 3% BSA in PBS, incubate cells with primary antibodies (1:500 dilution) for 1 hour at room temperature. After washing, incubate with fluorescently labeled secondary antibodies (1:1000) for 45 minutes, counterstain with DAPI, and mount [8].
Validation Criteria: The antibody is considered specific if:
- A band at the expected molecular weight (approximately 54-60 kDa) is present in WT but absent or dramatically reduced in KO lysates.
- The subcellular localization pattern (nuclear for NDR1, cytoplasmic for NDR2) is observed in WT but not KO cells.
- No cross-reactivity with the other NDR kinase is detected [8].
Reporter Design Strategies for Studying NDR Localization and Function
Fluorescent Protein Reporters for Live-Cell Imaging
Genetically encoded fluorescent reporters provide powerful tools for studying the dynamic localization and trafficking of NDR kinases in live cells. These approaches circumvent potential artifacts associated with fixation and immunostaining while enabling real-time observation of cellular processes.
Design Considerations: When creating NDR fluorescent reporters, several factors must be addressed:
- Tag Position: Both N- and C-terminal fusion proteins should be tested, as tag placement can affect protein folding, activity, and localization.
- Linker Length: Incorporating flexible linkers (e.g., GSG repeats) between the fluorescent protein and NDR kinase can minimize steric interference.
- Fluorescent Protein Selection: Photoswitchable or photoactivatable proteins (e.g., PA-GFP, Dronpa) enable advanced applications like tracking protein mobility and super-resolution imaging [31].
Verification Steps:
- Confirm that the tagged protein expresses at levels comparable to endogenous NDR.
- Verify that the fusion protein retains kinase activity and proper regulation.
- Validate localization patterns against validated antibodies in fixed cells [8].
Advanced Applications: Point-localization superresolution microscopy techniques, such as Photoactivation Localization Microscopy (PALM) and Stochastic Optical Reconstruction Microscopy (STORM), can achieve resolution down to tens of nanometers, allowing detailed visualization of NDR distribution within cellular compartments [31].
MRI-Based Reporter Systems
While fluorescent proteins are invaluable for cellular imaging, magnetic resonance imaging (MRI) reporters enable non-invasive detection of labeled cells in living animals. Recent research has identified effective reporter gene combinations for cellular-level MRI:
Transferrin Receptor: Facilitates cellular uptake of transferrin-bound iron and other metals like manganese [32].
Divalent Metal Transporter 1 (DMT1): Transports metal ions from transferrin receptor to intracellular destinations [32].
Ferritin-M6A: An engineered fusion protein combining ferritin with part of Mms6 from magnetic bacteria, demonstrating enhanced iron storage capacity compared to wild-type ferritin [32].
Studies have identified that the combination of transferrin receptor, DMT1, and Ferritin-M6A provides optimal contrast for T1-weighted imaging (T1WI), while Ferritin-M6A alone performs best for T2-weighted imaging (T2WI) [32]. These reporter systems could be adapted to study NDR kinase function in vivo, particularly in contexts where NDRs regulate endomembrane trafficking and transferrin receptor distribution [28].
Table 2: Comparison of Localization Reporter Strategies for NDR Kinases
| Method | Resolution | Applications | Advantages | Limitations |
|---|---|---|---|---|
| Immuno-fluorescence | ~200 nm (diffraction-limited) | Fixed cell imaging, subcellular localization | High specificity with validated antibodies; multiplexing capability | Requires cell fixation; potential antibody cross-reactivity |
| Genetically Encoded Fluorescent Proteins | ~200 nm (diffraction-limited) | Live-cell imaging, dynamics, trafficking | Enables real-time tracking in living cells; genetic targeting | Tag may affect protein function; photobleaching |
| PALM/STORM | ~20 nm | Nanoscale localization, molecular mapping | Ultra-high resolution; single-molecule sensitivity | Specialized equipment; complex sample preparation |
| MRI Reporters | ~100 μm (clinical) to cellular level | In vivo imaging, whole-organism studies | Non-invasive; deep tissue penetration; translational potential | Lower resolution; indirect detection of reporter |
Experimental Protocol: Live-Cell Imaging of NDR Kinase Trafficking
This protocol outlines the procedure for tracking NDR kinase localization and dynamics in live cells using fluorescent protein tags:
Plasmid Construction:
- Amplify NDR1 or NDR2 coding sequences from cDNA using high-fidelity PCR.
- Clone into mammalian expression vectors containing fluorescent proteins (e.g., EGFP, mCherry) using appropriate restriction sites or recombination cloning.
- Verify all constructs by sequencing before use [8].
Cell Transfection:
- Plate appropriate cells (e.g., COS-7, HEK293, or neuronal cells) on glass-bottom dishes 24 hours before transfection.
- Transfect with plasmid DNA using lipofection (e.g., Lipofectamine 3000) or electroporation according to manufacturer's instructions.
- Include controls expressing fluorescent protein alone to assess background localization [8].
Live-Cell Imaging:
- Image cells 24-48 hours post-transfection in live-cell imaging medium at 37°C with 5% CO2.
- For time-lapse imaging, acquire images at appropriate intervals (e.g., every 30 seconds to 5 minutes) depending on the process being studied.
- Use confocal or spinning disk microscopy for improved resolution of subcellular localization.
Image Analysis:
- Quantify nuclear-to-cytoplasmic ratio using image analysis software by defining regions of interest for nucleus and cytoplasm.
- For tracking intracellular movement, use particle tracking algorithms to analyze velocity, trajectory, and displacement.
- Correlate NDR localization with cellular compartments using organelle-specific markers [8].
Research Reagent Solutions for NDR Kinase Studies
Table 3: Essential Research Reagents for NDR Kinase Investigation
| Reagent Category | Specific Examples | Application/Function |
|---|---|---|
| Validated Antibodies | Anti-NDR1 (Santa Cruz, Transduction Labs), Anti-NDR2, Phospho-specific Anti-T444-P [8] | Western blot, immunofluorescence, immunoprecipitation |
| Cell Lines | HEK293, HeLa, U2OS, COS-7, Primary neurons [8] [28] | Biochemical studies, localization, functional assays |
| Knockout Models | NDR1 constitutive knockout mice, NDR2-floxed mice [26] [28] | In vivo functional studies, antibody validation |
| Expression Vectors | pcDNA3-NDR1/2, pEGFP-NDR1/2, CAG-promoter constructs [32] [8] | Ectopic expression, live-cell imaging |
| Kinase Assay Components | Active MST3 kinase, Okadaic acid, MOB proteins [27] [8] | In vitro kinase assays, pathway activation studies |
Signaling Pathways and Experimental Workflows
Experimental Workflow for NDR Localization Studies
NDR Kinase Signaling Pathways and Functional Outcomes
The precise differentiation between NDR1 and NDR2 through validated detection strategies reveals not merely technical requirements but fundamental biological insights into the functional specialization of these highly similar kinases. The distinct subcellular localization of NDR1 (nuclear) and NDR2 (cytoplasmic) underpins their specialized roles in critical cellular processes—from cell cycle regulation and antiviral defense to autophagy and membrane trafficking. The experimental frameworks and validation strategies presented here provide researchers with robust methodologies to advance our understanding of NDR kinase biology in both health and disease. As research continues to elucidate the complex signaling networks coordinated by these kinases, the implementation of rigorous detection and validation approaches will be paramount for generating reproducible, biologically relevant data with potential therapeutic implications for cancer, neurodegenerative disorders, and viral infections.
In modern molecular biology, selecting the appropriate genetic manipulation tool is paramount for designing rigorous and interpretable experiments. For researchers investigating nuanced biological questions, such as the functional differences between the highly similar kinases NDR1 (which displays nuclear localization) and NDR2 (primarily cytoplasmic), the choice of tool can define the project's success [8]. While all major tools aim to modulate gene function, their mechanisms, timelines, and resulting phenotypic readouts differ significantly.
This guide provides an objective comparison of three foundational technologies—CRISPR-Cas9 knockout, siRNA knockdown, and constitutively active kinase constructs—focusing on their performance in advanced cell biology research. We summarize quantitative data, detail experimental protocols, and visualize core concepts to equip scientists with the information needed to select the optimal tool for probing kinase function and subcellular localization.
Tool Comparison: Mechanisms and Applications
The table below summarizes the core characteristics of the three genetic manipulation tools, providing a direct comparison of their typical use cases, mechanisms, and experimental timelines.
Table 1: Comparison of Key Genetic Manipulation Tools
| Feature | CRISPR-Cas9 Knockout | siRNA Knockdown | Constitutively Active Kinase |
|---|---|---|---|
| Primary Goal | Permanent gene disruption | Temporary reduction of gene expression | Sustained pathway activation |
| Mechanism of Action | Creates double-strand breaks, leading to frameshift mutations and premature stop codons [33]. | Degrades target mRNA or inhibits its translation via the RNA-induced silencing complex (RISC) [34] [35]. | Mimics a kinase's active state, often via point mutations that prevent autoinhibition [8]. |
| Target Molecule | Genomic DNA | Messenger RNA (mRNA) | Protein function |
| Typical Timeline for Effect | Days to weeks (requires cell division and turnover) | Hours to days (effect diminishes with cell division and mRNA turnover) | Hours (after protein expression) |
| Permanence | Heritable, permanent change | Transient, reversible effect | Transient unless integrated into genome |
| Key Application in Kinase Research | Determining essential gene functions and long-term phenotypic consequences [36] [33]. | Studying acute gene function, validating drug targets, and probing essential genes [37]. | Investigating downstream signaling pathways and gain-of-function phenotypes [8]. |
Application in NDR Kinase Localization and Function Research
The case of human NDR kinases exemplifies how these tools can be deployed to dissect distinct biological questions. Studies show that while NDR1 and NDR2 are highly homologous, their subcellular localization differs, suggesting non-redundant functions [8]. The following diagram illustrates the workflow for applying these tools to study such a research problem.
Probing Localization with Constitutively Active Constructs
Research into the regulation of NDR kinase localization provides a classic example of using constitutively active constructs. One study demonstrated that membrane targeting of NDR kinases, facilitated by interaction with MOB proteins, resulted in their rapid activation through phosphorylation at specific sites (Ser281 and Thr444 for NDR1) [8]. This approach helped establish that the kinase's subcellular location is a critical regulator of its activity.
Key Experimental Protocol: Generating a Constitutively Active NDR Kinase
- Molecular Cloning: Fuse the kinase of interest (e.g., NDR1) to a proven membrane-targeting sequence, such as the myristoylation/palmitylation motif from the Lck tyrosine kinase (MGCVCSSN) [8].
- Cell Transfection & Culture: Transfect constructs into relevant cell lines (e.g., COS-7, U2-OS, HEK 293) using standard reagents like Fugene 6 or Lipofectamine 2000.
- Validation & Analysis:
- Biochemical: Use phospho-specific antibodies to confirm activation-state phosphorylation (e.g., anti-T444-P for NDR1) via immunoblotting [8].
- Cell Biological: Employ immunofluorescence and confocal microscopy to verify the forced re-localization (e.g., to the plasma membrane) and its effects.
Experimental Protocols and Workflows
CRISPR-Cas9 Knockout for In Vivo Screening
Traditional CRISPR screens in complex in vivo models like tumors are confounded by bottlenecks in cell engraftment and heterogeneous clonal growth. The novel CRISPR-StAR (Stochastic Activation by Recombination) method overcomes this by creating internal controls on a single-cell level [36].
Key Experimental Protocol: CRISPR-StAR Screen
- Library Design: Clone an sgRNA library (e.g., 5,870 sgRNAs targeting 1,245 genes) into the CRISPR-StAR backbone, which uses intercalated loxP/lox5171 sites for inducible sgRNA activation [36].
- Cell Engineering: Stably express Cas9 and Cre::ERT2 in the target cell line (e.g., mouse melanoma cells). Transduce the library at high coverage (>1,000 cells/sgRNA).
- In Vivo Screening & Analysis:
- Inject transduced cells into animal models and allow tumors to establish.
- Administer tamoxifen to induce Cre::ERT2, stochastically activating the sgRNA in a subset of cells within each clonal population. This creates isogenic internal controls (active sgRNA vs. inactive sgRNA) within the same tumor microenvironment [36].
- Harvest tumors, sequence the integrated barcodes (UMIs), and compare the abundance of active versus inactive sgRNAs within each clonal population to identify gene dependencies specific to the in vivo context.
The following diagram outlines the core logic of this advanced screening method.
siRNA Knockdown for Target Validation and Phenotypic Analysis
siRNA screens are powerful for identifying novel therapeutic targets. A screen investigating ciliopathies identified ROCK2 as a key mediator of cilium formation. This finding was validated by showing that ROCK inhibitors, like fasudil, could rescue ciliary function, suggesting a potential path for drug repurposing [37].
Key Experimental Protocol: Genome-Wide siRNA Reverse Genetics Screen
- Screen Design: Perform a whole-genome siRNA screen in a relevant cell model (e.g., hTERT RPE-1 or mouse IMCD-3 cells) to identify positive modulators of a specific phenotype, such as cilia formation [37].
- Phenotypic Analysis: Fix cells and stain for specific markers (e.g., anti-ARL13B or acetylated α-tubulin for cilia). Use high-content imaging (confocal microscopy) and automated image analysis (e.g., with FIJI software) to quantify phenotypes like cilia incidence and length [37].
- Hit Validation: Treat cells with pharmacological inhibitors (e.g., fasudil or the specific ROCK2 inhibitor belumosudil) to confirm that the phenotype observed with genetic knockdown can be recapitulated with chemical inhibition [37].
Technical Specifications and Optimization
Optimizing siRNA for Efficacy and Specificity
The efficacy of siRNA is not arbitrary; it depends on a set of well-defined sequence and structural features. Systematic studies have quantified the impact of these parameters on knockdown efficiency [34] [35].
Table 2: Key Design Parameters for Effective siRNA
| Parameter | Ineffective Design | Optimal Design | Impact on Efficacy |
|---|---|---|---|
| Length | < 17 nucleotides (nt) [34] | 19-21 nt [34] | Drastic loss of efficacy below 17 nt; 19 bp is standard. |
| Overhang Structure | Blunt ends [34] [35] | 2-nt 3' overhangs [34] [35] | SiRNAs with overhangs show enhanced efficacy compared to blunt-ended forms. |
| GC Content | ≥ 60% [35] | 30-50% [35] | High GC content is known to negatively impact silencing. |
| Target mRNA Context | Ignoring exon usage, polyadenylation sites, and ribosomal occupancy [35]. | Selecting target sites considering native mRNA structure and features [35]. | Native mRNA-specific features significantly influence siRNA performance. |
| Chemical Modification (2'-OMe) | Unmodified or suboptimal modification patterns [35]. | Specific, optimized patterns of 2'-O-methyl (2'-OMe) or 2'-fluoro (2'-F) modifications [35]. | The modification pattern significantly impacts efficacy and stability, while structural features (symmetric vs. asymmetric) have a lesser effect [35]. |
The Scientist's Toolkit: Essential Research Reagents
Successful execution of genetic manipulation experiments requires a suite of reliable reagents. The table below lists key materials and their functions as derived from the cited protocols.
Table 3: Essential Research Reagents for Genetic Manipulation Studies
| Reagent / Resource | Function / Application | Example Sources / Models |
|---|---|---|
| hTERT RPE-1 Cells | Non-cancerous, epithelial cell line ideal for ciliogenesis and cell biology studies [37]. | American Type Culture Collection (ATCC) |
| Anti-ARL13B Antibody | A high-quality primary antibody for immunofluorescence staining of primary cilia [37]. | Proteintech (17711-1-AP) |
| Fasudil Hydrochloride | A potent, broad ROCK inhibitor used to validate genetic findings related to ROCK2 and cilia function [37]. | Generic, off-patent drug |
| CRISPR-StAR Vector | A specialized plasmid for inducible, internally controlled CRISPR screening in complex in vivo models [36]. | Custom cloning |
| Lipofectamine 2000 | A common transfection reagent for delivering nucleic acids (plasmids, siRNA) into a variety of cell lines [8]. | Thermo Fisher Scientific |
| Phospho-specific Antibodies | Critical reagents for detecting the activated, phosphorylated state of kinases (e.g., anti-T444-P for NDR1) [8]. | Custom generation or commercial (e.g., Cell Signaling Technology) |
| Okadaic Acid (OA) | A PP2A phosphatase inhibitor used to study kinase activation pathways, as in NDR kinase research [8]. | Alexis Corp. (Enzo Life Sciences) |
| C-021 | CCR4 Antagonist C-021|Research Compound | |
| Isrib | ISRIB|Integrated Stress Response Inhibitor|eIF2B Activator | ISRIB is a potent small molecule inhibitor of the integrated stress response (ISR) that reverses the effects of eIF2α phosphorylation. This product is For Research Use Only. Not for human or veterinary diagnostic or therapeutic use. |
The Nuclear Dbf2-related (NDR) serine/threonine kinases, NDR1 and NDR2, represent a fascinating paradigm in cellular signaling where highly similar proteins (sharing approximately 87% amino acid identity) exhibit distinct subcellular localizations that dictate their specialized biological functions [6]. Despite their significant structural similarity, these kinases display remarkably different distribution patterns within cells: NDR1 predominantly localizes to the nucleus, while NDR2 exhibits a punctate cytoplasmic distribution and associates with membrane compartments, including peroxisomes [4] [6]. This differential localization is not merely incidental but appears crucial for their specialized roles in cellular processes ranging from centrosome duplication and mitotic chromosome alignment to ciliogenesis and membrane trafficking [4] [28].
The functional consequences of this compartmentalization are significant. NDR2's specific peroxisomal localization, mediated by its C-terminal Gly-Lys-Leu (GKL) motif functioning as a peroxisome-targeting signal, is essential for its role in promoting primary cilium formation [4]. In contrast, NDR1's nuclear presence implicates it in distinct regulatory pathways. These localization-dependent functions underscore the critical importance of precise subcellular fractionation techniques for researchers investigating the unique roles of these kinases in normal cellular physiology and disease states, including cancer and neurodegenerative conditions [28] [38]. This guide provides a comprehensive comparison of methodologies for isolating and studying these spatially distinct kinase pools.
Key Principles of Subcellular Fractionation
Subcellular fractionation relies on the fundamental principle that cellular organelles and compartments possess distinct physical properties, including size, shape, density, and biochemical composition [39] [40]. By exploiting these differences through carefully optimized centrifugation techniques and buffer systems, researchers can separate cellular components with sufficient purity for downstream analysis. The general workflow involves two primary approaches: differential centrifugation, which separates components based on sedimentation rate largely determined by size, and density gradient centrifugation, which provides higher purity separation based on buoyant density [39].
For NDR kinase studies, maintaining protein activity and post-translational modification states is paramount, requiring careful attention to protease and phosphatase inhibition throughout the process [40]. The divergent localizations of NDR1 and NDR2 present a unique challenge, necessitating protocols that can effectively separate nuclear, cytoplasmic, and membrane compartments while preserving the integrity of kinase complexes and their activation states, which are regulated by phosphorylation and interaction with MOB proteins [17] [8].
Experimental Protocols for NDR Kinase Fractionation
Sequential Differential Centrifugation Protocol
The following protocol, adapted from established methodologies [41] [40], provides a robust foundation for separating nuclear, cytoplasmic, and membrane-associated NDR kinases:
Cell Lysis Buffer Preparation: 20 mM HEPES (pH 7.4), 10 mM KCl, 2 mM MgClâ‚‚, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and fresh protease/phosphatase inhibitors. For membrane protein studies, 0.1% digitonin or Triton X-114 can be added for initial permeabilization [42] [40].
Stepwise Procedure:
- Cell Harvesting: Culture cells in 10 cm plates, wash with ice-cold PBS, and scrape into 500 μL fractionation buffer.
- Permeabilization: Incubate cell suspension on ice for 15 minutes. Pass through a 27-gauge needle 10 times. Incubate again on ice for 20 minutes.
- Nuclear Fraction: Centrifuge at 720 × g for 5 minutes at 4°C. The pellet contains the nuclei. Wash pellet with 500 μL fractionation buffer, disperse by pipetting, and pass through a 25-gauge needle 10 times. Centrifuge again at 720 × g for 10 minutes. Resuspend the final nuclear pellet in TBS with 0.1% SDS and briefly sonicate to shear genomic DNA.
- Organellar/Membrane Fraction: Transfer the post-nuclear supernatant to a fresh tube and centrifuge at 10,000 × g for 5 minutes. The pellet contains mitochondria and other large organelles. For NDR2 studies, this fraction will contain a significant portion of peroxisome-associated kinase.
- Membrane Fraction: Transfer the supernatant from step 4 to an ultracentrifuge tube and centrifuge at 100,000 × g for 1 hour. The resulting pellet contains membrane vesicles and associated proteins, including membrane-bound NDR kinases.
- Cytosolic Fraction: The supernatant from step 5 represents the soluble cytosolic fraction, which can be concentrated using centrifugal filter units if necessary.
Quality Control: Assess fraction purity by Western blotting using compartment-specific markers [40]: Lamin A/C (nucleus), Catalase (peroxisomes), TOMM20 (mitochondria), Calnexin (endoplasmic reticulum), and Tubulin (cytosol).
Density Gradient Purification for Peroxisomal NDR2
For studies specifically focusing on NDR2's peroxisomal localization, density gradient centrifugation provides superior purity [4]:
- Prepare a discontinuous iodixanol gradient (e.g., 17%, 25%, 35%, 50%) in an ultracentrifuge tube.
- Layer the post-nuclear supernatant (or the 10,000 × g pellet resuspended in appropriate buffer) carefully on top of the gradient.
- Centrifuge at >100,000 × g for 2-3 hours at 4°C.
- Collect distinct bands corresponding to different organelles using a Pasteur pipette. Peroxisomes typically band at higher densities (approximately 1.18 g/cm³ in iodixanol).
- Verify peroxisomal fractions by Western blot for catalase and Pex14p [4].
The following diagram illustrates the core workflow for separating NDR1 and NDR2 pools:
Comparative Analysis of Fractionation Techniques
Technical Comparison of Fractionation Methods
Table 1: Comparison of Subcellular Fractionation Methods for NDR Kinase Studies
| Method | Principle | Purity/Yield | NDR1 Recovery | NDR2 Recovery | Downstream Applications | Time Requirement |
|---|---|---|---|---|---|---|
| Differential Centrifugation | Sequential centrifugation at increasing speeds | Moderate purity, high yield | Effective in nuclear fraction | Effective in membrane/organelle fractions | Western blot, activity assays, protein interaction studies | 2-3 hours |
| Density Gradient Centrifugation | Separation by buoyant density in sucrose/iodixanol | High purity, moderate yield | Limited utility | Excellent for peroxisomal NDR2 isolation [4] | Proteomics, precise localization studies, functional analysis of purified organelles | 4-6 hours (including ultracentrifugation) |
| Commercial Kit Systems | Optimized reagent-based separation | Consistent purity, variable yield | Good for nuclear extracts | Effective for cytoplasmic/membrane partitioning [42] [41] | Rapid screening, standardized comparisons, lower-throughput studies | 1-2 hours |
Quantitative Distribution of NDR Kinases Across Fractions
Table 2: Expected Distribution Patterns of NDR1 and NDR2 in Subcellular Fractions
| Subcellular Fraction | Primary Marker | NDR1 Localization | NDR2 Localization | Functional Significance |
|---|---|---|---|---|
| Nuclear | Lamin A/C | Strong (predominant) [6] | Weak/Absent [6] | Cell cycle regulation, transcription control [4] |
| Cytosolic | Tubulin | Moderate | Moderate (diffuse) | Kinase activation, MOB protein interactions [8] |
| Membrane | Calnexin | Weak | Strong (punctate) [6] | Signal transduction, membrane trafficking [28] |
| Peroxisomal | Catalase/Pex14p | Absent | Strong (C-terminal GKL dependent) [4] | Ciliogenesis regulation [4] |
| Mitochondrial | TOMM20 | Weak/Moderate | Weak/Moderate | Apoptosis regulation, metabolic signaling [28] |
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Reagents for NDR Kinase Fractionation and Analysis
| Reagent/Category | Specific Examples | Function in NDR Studies |
|---|---|---|
| Fractionation Kits | Thermo Scientific Subcellular Protein Fractionation Kit [42]; Abcam Cell Fractionation Kits [41] | Standardized protocols for reproducible separation of nuclear, cytoplasmic, and membrane compartments |
| Centrifugation Media | Sucrose; Iodixanol; Percoll | Density gradient formation for high-purity organelle separation, particularly peroxisomal NDR2 [4] |
| Protease Inhibitors | PMSF; Leupeptin; Aprotinin | Prevent protein degradation during fractionation |
| Phosphatase Inhibitors | NaF; β-glycerol phosphate; Na₃VO₄ | Preserve phosphorylation states at critical sites (NDR1: Ser281/Thr444; NDR2: Ser282/Thr442) [17] [8] |
| Detergents | Digitonin; Triton X-114; Non-idet P-40 | Selective membrane permeabilization and protein extraction |
| Validation Antibodies | Anti-NDR1 (specific); Anti-NDR2 (C-terminal specific) [38]; Phospho-specific NDR (Ser281/282, Thr444/442) [17] [8] | Detection and discrimination between NDR isoforms and their activation states |
| Compartment Markers | Lamin A/C (nuclear); Catalase (peroxisomal) [4]; Tubulin (cytosolic); Calnexin (ER) | Assessment of fraction purity and cross-contamination |
| W123 | W123, MF:C17H26N2O3, MW:306.4 g/mol | Chemical Reagent |
| OU749 | OU749 CAS 519170-13-9|GGT Inhibitor | OU749 is a non-glutamine, uncompetitive, and species-specific GGT inhibitor for research. For Research Use Only. Not for human use. |
Troubleshooting and Quality Control
Successful fractionation requires rigorous quality control. Common challenges include:
- Cross-contamination between fractions: Mitigate by optimizing centrifugation conditions and using gradient purification for critical applications. Always validate with compartment-specific markers.
- Protein degradation: Maintain samples at 4°C throughout the process and use fresh protease inhibitors.
- Loss of post-translational modifications: Include appropriate phosphatase inhibitors and avoid repeated freeze-thaw cycles.
- NDR2 peroxisomal localization verification: Confirm using catalase co-localization and demonstrate dependence on the C-terminal GKL motif [4].
The following diagram illustrates the regulatory network governing NDR kinase activation and their distinct downstream functions, highlighting the importance of precise subcellular localization:
The distinct subcellular localizations of NDR1 and NDR2 kinases represent a crucial determinant of their specialized functions in cellular regulation and disease pathogenesis. The fractionation protocols detailed in this guide provide essential methodologies for researchers to isolate and characterize these spatially segregated kinase pools. As research continues to elucidate the complex roles of NDR kinases in processes ranging from neuronal health to cancer biology, precise subcellular fractionation will remain an indispensable tool for understanding the compartment-specific signaling networks that govern cellular behavior. The experimental approaches outlined here offer a foundation for investigating how these highly similar kinases achieve their functional specificity through strategic partitioning within the cellular architecture.
The Nuclear Dbf2-related (NDR) kinases, NDR1 (STK38) and NDR2 (STK38L), are serine/threonine kinases belonging to the NDR/LATS subfamily of the AGC kinase group and are highly conserved from yeast to humans [43] [3]. Despite their high sequence similarity (approximately 87% identity), they exhibit distinct subcellular localization patterns that dictate their specialized biological functions. NDR1 is predominantly localized in the nucleus, while NDR2 displays a cytoplasmic distribution and exhibits unique punctate localization patterns [4] [3]. This fundamental difference in localization is critical for understanding their respective roles in transcriptional regulation and DNA damage response (for NDR1) versus vesicle trafficking, autophagy, and ciliogenesis (for NDR2) [4] [44] [45].
The regulatory mechanism of NDR kinases involves phosphorylation and interaction with MOB (Mps1 One Binder) proteins. Both NDR1 and NDR2 require phosphorylation on specific sites (Ser281/Ser282 in the T-loop and Thr444/Thr442 in the hydrophobic motif) for full activation [8]. Membrane targeting of NDR kinases results in constitutive activation that can be further enhanced by co-expression of MOB proteins [8]. This review will focus specifically on functional assays for NDR1, with an emphasis on its nuclear functions in transcriptional regulation and DNA damage response, while providing comparative insights into NDR2's cytoplasmic roles.
NDR1 in Transcriptional Regulation: Assays and Mechanistic Insights
NDR1 plays a significant role in transcriptional regulation through both Hippo pathway-dependent and independent mechanisms. As part of the Hippo signaling cascade, NDR1, along with NDR2, functions as an upstream kinase of the transcriptional co-activators YAP and TAZ [43] [46]. In its active state, the Hippo pathway leads to phosphorylation and cytoplasmic sequestration of YAP/TAZ, thereby inhibiting their transcriptional activity.
Hippo Pathway-Dependent Transcriptional Regulation
Experimental Protocol: YAP/TAZ Localization and Phosphorylation Assay
- Cell Culture and Transfection: Culture HEK293 or relevant cell lines and transfect with NDR1-specific siRNA or overexpression constructs.
- Stimulation and Fixation: After 48-72 hours, stimulate cells as needed and fix with 4% paraformaldehyde.
- Immunofluorescence Staining: Incubate cells with anti-YAP/TAZ primary antibodies followed by fluorescently-labeled secondary antibodies. Use DAPI for nuclear counterstaining.
- Imaging and Analysis: Visualize using confocal microscopy. Quantify nuclear-to-cytoplasmic YAP/TAZ ratio across multiple cells.
- Western Blot Validation: Analyze cell lysates using phospho-specific YAP antibodies (e.g., Ser127) to confirm phosphorylation status.
Key Research Reagents:
- NDR1 siRNA: Silences NDR1 expression to assess loss-of-function phenotypes.
- Anti-YAP/TAZ Antibodies: Detect subcellular localization of these transcriptional co-activators.
- Phospho-Specific YAP Antibodies: Monitor Hippo pathway activity through phosphorylation status.
- Luciferase Reporter Constructs: Measure TEAD transcriptional activity downstream of YAP/TAZ.
Hippo Pathway-Independent Transcriptional Regulation
NDR1 also regulates transcription through non-Hippo mechanisms. It binds to the intergenic region of miR146a, dampening its transcription independently of its kinase activity [3]. This regulation subsequently promotes STAT1 translation, enhancing type I interferon production and antiviral immune responses.
NDR1 in DNA Damage Response: Functional Assays and Protocols
NDR1 plays a critical role in the cellular response to DNA damage, particularly in the nucleotide excision repair (NER) pathway that addresses UV-induced DNA lesions [44]. Upon UV irradiation, NDR1 accumulates in the nucleus and interacts with XPA (xeroderma pigmentosum A), a rate-limiting factor in NER that helps verify DNA damage.
NDR1-XPA Interaction and UV-Induced Nuclear Accumulation
Experimental Protocol: NDR1 Nuclear Translocation Assay
- UV Treatment: Culture quiescent cells and irradiate with UV-C (10-20 J/m²) using a UV crosslinker.
- Time-Course Analysis: Harvest cells at various time points post-irradiation (0, 15, 30, 60, 120 min).
- Subcellular Fractionation: Lyse cells and separate nuclear and cytoplasmic fractions using differential centrifugation.
- Western Blot Analysis: Probe fractions with anti-NDR1, anti-lamin A/C (nuclear marker), and anti-α-tubulin (cytoplasmic marker) antibodies.
- Quantification: Normalize NDR1 levels in nuclear fractions to lamin A/C across time points.
Experimental Protocol: Co-Immunoprecipitation of NDR1-XPA Complex
- Cell Lysis: Harvest UV-irradiated cells using non-denaturing lysis buffer.
- Immunoprecipitation: Incubate lysates with anti-NDR1 or control IgG antibodies overnight at 4°C.
- Pull-Down: Add protein A/G beads, incubate, and wash extensively.
- Detection: Elute proteins, separate by SDS-PAGE, and immunoblot with anti-XPA and anti-NDR1 antibodies.
Functional Impact on DNA Repair Efficiency
Experimental Protocol: Cyclobutane Pyrimidine Dimer (CPD) Repair Assay
- UV Irradiation and Recovery: Treat cells with UV (10-20 J/m²) and allow recovery for various durations (0-24 hours).
- DNA Extraction: Isolate genomic DNA at each time point.
- CPD Quantification: Use ELISA with anti-CPD antibodies or slot-blot analysis to measure CPD levels.
- Data Analysis: Calculate repair kinetics by fitting CPD removal data to exponential decay curves.
The knockdown of NDR1 delays the repair of UV-induced cyclobutane pyrimidine dimers without altering the expression levels or chromatin association of core NER factors [44]. Instead, NDR1-depleted cells show reduced ATR kinase activity toward specific substrates including CHK1 and p53, indicating that NDR1 modulates NER indirectly via the ATR pathway.
Comparative Functional Analysis: NDR1 versus NDR2
Table 1: Comparative Analysis of NDR1 and NDR2 Functions and Assays
| Feature | NDR1 (STK38) | NDR2 (STK38L) |
|---|---|---|
| Primary Localization | Nuclear [3] | Cytoplasmic, peroxisomal [4] |
| Transcriptional Role | Direct YAP phosphorylation; miR146a regulation [43] [3] | YAP phosphorylation; limited nuclear role [43] |
| DNA Damage Response | UV-induced nuclear accumulation; XPA interaction; ATR modulation [44] | Not established |
| Specialized Functions | Cell cycle regulation (G1/S, G2/M) [46] | Ciliogenesis via Rabin8 phosphorylation; vesicle trafficking [4] |
| Key Functional Assays | YAP localization, XPA co-IP, CPD repair, nuclear translocation | Rabin8 phosphorylation, ciliogenesis scoring, peroxisomal targeting |
Table 2: Quantitative Data from Key NDR1 Functional Studies
| Experimental Readout | Control Condition | NDR1 Manipulation | Biological System | Reference |
|---|---|---|---|---|
| CPD Repair Half-time | ~4-6 hours | Increased by ~50-100% with NDR1 knockdown | Human cell lines | [44] |
| YAP Nuclear/Cytoplasmic Ratio | ~0.3 (phospho-mimetic active NDR1) | ~1.2 (kinase-dead NDR1) | HEK293 cells | [43] |
| UV-Induced Nuclear Accumulation | 2.5-fold increase at 60 min post-UV | No significant nuclear accumulation in NDR1 KD | Quiescent fibroblasts | [44] |
| IL-6 Production (CpG-induced) | Baseline (100%) | Increased by ~40-60% in NDR1 KO | Macrophages, mouse model | [3] |
Signaling Pathways and Experimental Workflows
Diagram 1: NDR1 Signaling in Transcriptional Regulation and DNA Damage Response
Diagram 2: Experimental Workflow for NDR1 DNA Damage Response Assays
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for NDR1 Functional Studies
| Reagent Category | Specific Examples | Function/Application | Experimental Notes |
|---|---|---|---|
| Cell Lines | HEK293, RPE1, Quiescent fibroblasts | General NDR1 functional studies; DNA damage response | RPE1 useful for localization studies; primary for damage response |
| Antibodies | Anti-NDR1 (specific C-terminal), Anti-phospho-NDR1 (Thr444/Ser281), Anti-XPA, Anti-YAP/TAZ, Anti-phospho-YAP (Ser127) | Detection, localization, phosphorylation status, interaction studies | Validate species specificity; phospho-antibodies require peptide competition |
| Molecular Tools | NDR1 siRNA/shRNA, Kinase-dead NDR1 (K118A), Constitutively active NDR1, NDR1-GFP fusion | Loss/gain-of-function, localization tracking, mechanistic studies | Monitor off-target effects with siRNA; verify expression with rescue experiments |
| Specialized Assays | CPD ELISA kit, Subcellular fractionation kit, Co-IP reagents, Luciferase reporter constructs (TEAD) | DNA repair quantification, nuclear translocation, protein interactions, transcriptional readout | Include proper controls for normalization (e.g., DAPI for nuclear counts) |
| Activation Methods | Okadaic acid (PP2A inhibitor), UV-C irradiation, MOB1 co-expression | NDR1 pathway activation, DNA damage induction | Optimize concentrations and time courses for specific applications |
| TMRM | TMRM, MF:C25H25N2O3+, MW:401.5 g/mol | Chemical Reagent | Bench Chemicals |
| 2-Acetyl-4-tetrahydroxybutyl imidazole | 2-Acetyl-4-tetrahydroxybutyl imidazole, CAS:94944-70-4, MF:C9H14N2O5, MW:230.22 g/mol | Chemical Reagent | Bench Chemicals |
The functional assays for NDR1 highlight its unique nuclear roles in transcriptional regulation and DNA damage response, distinguishing it from its cytoplasmic counterpart NDR2. The distinct subcellular localization of these highly similar kinases enables specialized cellular functions while maintaining some overlapping roles in Hippo signaling. The experimental approaches outlined here—including YAP/TAZ localization assays, UV-induced nuclear translocation studies, XPA interaction analyses, and DNA repair efficiency measurements—provide researchers with robust methodologies to investigate NDR1-specific functions.
Understanding these distinct roles has significant implications for drug development, particularly in cancer therapeutics and treatments for DNA repair deficiency disorders. The differential localization and function of NDR1 and NDR2 suggest they may represent distinct therapeutic targets, with NDR1 modulation potentially affecting genomic stability and transcriptional regulation, while NDR2 targeting might impact ciliogenesis-related pathologies and vesicular trafficking. Future research should focus on developing more specific inhibitors and activators that can distinguish between these two kinase isoforms, potentially leading to more targeted therapeutic interventions with reduced off-target effects.
The NDR (Nuclear Dbf2-related) serine-threonine kinase family, comprising NDR1 and NDR2, represents crucial regulators of cellular processes despite their high sequence similarity (approximately 87%). A fundamental distinction dictates their functional specialization: subcellular localization. NDR1 is characterized by nuclear distribution, while NDR2 exhibits a distinct punctate cytoplasmic pattern [6] [1]. This differential positioning suggests non-overlapping biological roles, with cytoplasmic NDR2 uniquely positioned to regulate processes including integrin signaling, vesicular trafficking, phagocytosis, and cell migration [47] [4] [12]. This guide provides a comparative analysis of functional assays for investigating NDR2-specific roles within the cytoplasm, offering experimental data and methodologies to elucidate its unique contributions to cellular physiology and disease contexts.
Functional Differences Between NDR1 and NDR2
Table 1: Fundamental Characteristics of NDR1 and NDR2
| Feature | NDR1 (STK38) | NDR2 (STK38L) |
|---|---|---|
| Subcellular Localization | Nuclear and diffuse cytoplasmic [6] [1] | Punctate cytoplasmic; peroxisomal localization [4] [6] |
| Tissue Expression | Widely expressed [1] | Highest expression in thymus; expressed in most tissues [1] |
| Peroxisomal Targeting Signal | Absent (C-terminal: Ala-Lys) [4] | Present (C-terminal: Gly-Lys-Leu) [4] |
| Role in Ciliogenesis | Non-essential [4] | Critical promoter [4] |
| Role in Neurite Outgrowth | Modulates dendritic growth and spine density [47] | Controls integrin-dependent dendritic and axonal growth [47] |
| Microglial Metabolic Adaptation | Not implicated in high-glucose response [12] | Key regulator under high-glucose conditions [12] |
The C-terminal peroxisome-targeting signal type 1 (PTS1) in NDR2 (Gly-Lys-Leu) facilitates its unique peroxisomal association, while NDR1 (C-terminal: Ala-Lys) lacks this signal [4]. This localization is functionally significant, as wild-type NDR2, but not a peroxisome-non-targeting mutant (NDR2(ΔL)), rescues ciliogenesis defects in NDR2-deficient cells [4].
NDR2-Specific Cytoplasmic Functional Assays
Phagocytosis and Migration Assays in Microglia
Table 2: Quantitative Functional Data from Microglial NDR2 Studies
| Assay Type | Experimental System | Key Finding with NDR2 Downregulation | Measurement Data |
|---|---|---|---|
| Phagocytic Capacity | BV-2 microglial cells (Ndr2 knockdown) | Significant reduction in phagocytosis [12] | Decreased uptake of phagocytic substrates |
| Migratory Capacity | BV-2 microglial cells (Ndr2 knockdown) | Significant impairment of migration [12] | Reduced migration in response to chemoattractants |
| Mitochondrial Respiration | BV-2 microglial cells (Ndr2 knockdown) | Impaired mitochondrial function and reduced metabolic flexibility [12] | Decreased oxygen consumption rate (OCR) |
| Cytokine Secretion | BV-2 microglial cells (Ndr2 knockdown) | Elevated pro-inflammatory cytokines (IL-6, TNF, IL-17, IL-12p70) [12] | Increased cytokine levels even under normal glucose |
| NDR2 Protein Expression | BV-2 cells under high glucose (30.5 mM) | Significant upregulation of NDR2 protein [12] | 7h assay: CT: 24.0 ± 4.4 a.u.; HG: 83.0 ± 19.1 a.u.12h assay: CT: 26.1 ± 6.9 a.u.; HG: 64.2 ± 10.1 a.u. |
Experimental Protocol: Microglial Phagocytosis and Migration Assays
- Cell Models: BV-2 immortalized microglial cells, primary mouse retinal microglial cultures, or human iPSC-derived microglial cells [12].
- NDR2 Manipulation: Implement CRISPR-Cas9-mediated knockdown using sgRNA targeting exon 7 of the Ndr2 gene or specific shRNA constructs [12].
- Phagocytosis Assay: Incubate cells with fluorescently labeled phagocytic substrates (E. coli particles, zymosan, or apoptotic cells). Quantify internalized particles using flow cytometry or fluorescence microscopy after trypan blue quenching of external fluorescence [12].
- Migration Assay: Perform transwell migration assays. Seed NDR2-manipulated microglia in upper chambers and measure movement toward chemoattractants (e.g., C5a, ATP) in lower chambers. Fixed cells are stained and quantified microscopically [12].
- High-Glucose Conditions: Expose cells to 30.5 mM glucose for 7-12 hours to mimic diabetic stress. Include osmotic controls (e.g., mannitol) to distinguish hyperglycemia-specific effects [12] [48].
Integrin Signaling and Neurite Outgrowth Assays
Experimental Protocol: Integrin-Dependent Neurite Outgrowth
- Cell Culture: Primary mouse hippocampal neurons [47].
- NDR2 Manipulation: Implement Ndr2 shRNA knockdown or use neurons from Ndr2-null mutant mice. Include rescue experiments with shRNA-resistant EGFP-Ndr2 constructs [47].
- Neurite Outgrowth Analysis: Transferd neurons with fluorescent markers (e.g., pCMV-tdTomato). Capture images at various developmental stages and quantify total dendritic length, branching complexity (Sholl analysis), and axonal length using image analysis software [47].
- Integrin Activation Assessment: Measure surface expression of activated β1-integrin using 9EG7 antibody via immunocytochemistry or flow cytometry. Alternatively, detect phosphorylation at Thr788/789 of β1-integrin using phospho-specific antibodies [47].
- Integrin Trafficking Assay: Co-transfect neurons with EGFP-Ndr2 and markers for early endosomes (Rab5) or recycling endosomes (Rab11). Analyze colocalization using confocal microscopy and quantitative image analysis [47].
Diagram 1: NDR2 in integrin signaling and neurite outgrowth (5 nodes)
Ciliogenesis Assays and Peroxisomal Localization
Experimental Protocol: Primary Cilium Formation and Peroxisome Association
- Cell Models: Human telomerase-immortalized retinal pigment epithelial (RPE1) cells or HeLa cells [4].
- Transfection and Staining: Transfect cells with YFP-NDR2 and peroxisomal markers (CFP-SKL or catalase antibodies). For endogenous detection, use antibodies against NDR2 and peroxisomal proteins (Pex14p, catalase) [4].
- Ciliogenesis Assay: Induce ciliogenesis by serum starvation for 24-48 hours. Fix cells and immunostain for ciliary markers (e.g., acetylated α-tubulin, Arl13b). Quantify ciliation percentage and cilium length using fluorescence microscopy [4].
- Subcellular Fractionation: Prepare post-nuclear supernatant fractions and separate using iodixanol density gradient ultracentrifugation. Analyze fractions by Western blotting for NDR2 and organelle markers [4].
- Functional Rescue: Express wild-type NDR2 and peroxisome-targeting-deficient mutant (NDR2(ΔL)) in NDR2-knockdown cells and assess ciliogenesis rescue capability [4].
Key Signaling Pathways and Molecular Mechanisms
NDR2-Mediated Integrin Activation Pathway
Molecular Mechanism: NDR2 stimulates integrin-dependent processes through phosphorylation at the activity- and trafficking-relevant site Thr788/789 of β1-integrin [47]. This promotes PKC- and CaMKII-dependent activation of β1-integrins and stimulates their exocytosis [47]. NDR2 associates with integrin-positive early and recycling endosomes in primary hippocampal neurons, regulating integrin trafficking to the cell surface [47].
Functional Outcome: This pathway is crucial for dendritic and axonal growth in mouse hippocampal neurons. Ndr2-null mutant mice exhibit arbor-specific alterations of dendritic complexity in the hippocampus, indicating a role in fine regulation of dendritic growth [47].
NDR2 in Metabolic Adaptation and Inflammatory Response
Molecular Mechanism: Under high-glucose conditions, NDR2 protein expression is significantly upregulated in microglial cells, regulating mitochondrial metabolism and cytoskeletal dynamics [12]. NDR2 downregulation impairs mitochondrial respiration, reduces metabolic flexibility, and elevates pro-inflammatory cytokines (IL-6, TNF, IL-17, IL-12p70) [12].
Functional Outcome: These mechanisms explain how NDR2 regulates microglial phagocytosis, migration, and inflammatory responses under diabetic conditions, contributing to neuroinflammatory processes in diabetic retinopathy [12].
Diagram 2: NDR2 in microglial dysfunction under high glucose (7 nodes)
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for NDR2 Functional Studies
| Reagent Category | Specific Examples | Research Application | Function in Assays |
|---|---|---|---|
| NDR2 Expression Constructs | EGFP-Ndr2, mCherry-Ndr2, shRNA-resistant EGFP-Ndr2 [47] | Localization, rescue experiments | Fluorescent tracking, functional complementation |
| NDR2 Mutants | Constitutively active Ndr2, NDR2(ΔL) peroxisomal mutant [47] [4] | Mechanism of action studies | Dissecting signaling requirements and localization |
| Knockdown Tools | shRNA against murine Ndr2, CRISPR-Cas9 with sgRNA targeting exon 7 [47] [12] | Loss-of-function studies | Investigating NDR2 deficiency effects |
| Antibodies for Detection | Anti-NDR2 (C-terminal), anti-pT788/789 β1-integrin, 9EG7 (activated β1-integrin) [47] [12] | Protein localization and activation | Immunofluorescence, Western blot, flow cytometry |
| Organelle Markers | CFP-SKL (peroxisomes), Rab5 (early endosomes), Rab11 (recycling endosomes) [47] [4] | Subcellular localization | Colocalization studies with NDR2 |
| Cell Type-Specific Markers | IBA1 (microglia), MAP2 (neurons), acetylated α-tubulin (cilia) [47] [12] | Cell identification and characterization | Validating cellular models and phenotypes |
| A 779 | A 779, MF:C39H60N12O11, MW:873.0 g/mol | Chemical Reagent | Bench Chemicals |
| AEM1 | AEM1|NRF2 Inhibitor | AEM1 is a potent NRF2 inhibitor with anti-tumor activity and oral efficacy. It sensitizes cancer cells to chemo. For Research Use Only. Not for human use. | Bench Chemicals |
The distinct cytoplasmic localization of NDR2, contrasted with NDR1's nuclear predominance, establishes its unique functional repertoire in regulating phagocytosis, migration, and integrin signaling. The experimental approaches detailed herein provide researchers with comprehensive methodologies to investigate NDR2-specific functions, with particular relevance to neurological development, microglial pathophysiology, and metabolic disease contexts. The continuing elucidation of NDR2-specific interactomes and signaling modules promises to reveal novel therapeutic targets for conditions including diabetic retinopathy, neurodegenerative disorders, and cancer metastasis [12] [10].
The nuclear Dbf2-related (NDR) kinases NDR1 (STK38) and NDR2 (STK38L) are serine/threonine kinases belonging to the NDR/LATS subfamily of the AGC protein kinase group, highly conserved from yeast to humans [3] [5]. Despite their high sequence similarity (approximately 87% identity), they exhibit fundamentally different subcellular localizations: NDR1 is predominantly nuclear, while NDR2 displays a punctate cytoplasmic distribution [1] [8]. This distinct compartmentalization suggests unique biological functions for each kinase, positioning them as key regulators in cellular processes ranging from morphogenesis and proliferation to apoptosis and immune response [3] [5]. Research into the NDR1/2 functional dichotomy relies heavily on specific model systems that can elucidate their context-dependent roles, particularly in specialized cells like neurons, microglia, and macrophages. This guide objectively compares the primary model systems used in NDR kinase research, evaluating their application in uncovering the distinct functions of nuclear NDR1 versus cytoplasmic NDR2 within the neuroimmune context.
Comparative Analysis of Model Systems in NDR Kinase Research
Table 1: Comparison of Key Model Systems for NDR1/2 Functional Studies
| Model System | Key Applications in NDR1/2 Research | Advantages | Limitations | Representative Findings |
|---|---|---|---|---|
| Immortalized Cell Lines (HeLa, HEK293, COS-7) | - Subcellular localization studies [1] [8]- Kinase activation mechanisms [8]- Protein-protein interactions | - High transfection efficiency- Reproducible results- Cost-effective maintenance | - Does not fully represent specialized primary cell physiology | - Identification of differential NDR1/2 localization [1]- MOB-dependent kinase activation at plasma membrane [8] |
| Primary Murine Macrophages & Microglia | - Innate immune response regulation [3]- Cytokine production studies- PRR signaling pathways (e.g., TLR9) | - Physiologically relevant immune responses | - Technically challenging isolation | - NDR1 negatively regulates TLR9-mediated inflammation via MEKK2 degradation [3] |
| Gene-Targeted Mouse Models (Knockout/Knockin) | - In vivo validation of NDR1/2 functions- Whole-organism physiology and development- Neuroimmune axis studies | - Provides complete physiological context | - High cost and time investment | - Stk38-deficient mice show heightened susceptibility to sepsis and E. coli infection [3] |
| Advanced 3D & Organ-on-a-Chip Models | - Neuroimmune interactions in AD/PD [49]- Complex cell-cell signaling- Therapeutic screening | - Recapitulates tissue-like architecture | - Technologically complex | - Potential for modeling NDR kinase roles in neuroinflammation and polarized cell signaling [49] |
Table 2: Quantitative Data from Key NDR1/2 Experimental Studies
| Experimental Context | Key Measured Parameters | NDR1-Specific Effects | NDR2-Specific Effects | Citation |
|---|---|---|---|---|
| TLR9 Signaling (CpG DNA) | TNF-α and IL-6 production | Significant increase in cytokines upon NDR1 deficiency [3] | Increased IL-6 secretion upon siRNA knockdown [3] | [3] |
| RIG-I Antiviral Signaling | Type I IFN and ISG production | Positively regulates via miR146a/STAT1 axis [3] | Promotes RIG-I/TRIM25 complex formation and K63 ubiquitination [3] | [3] |
| Subcellular Localization | Primary localization in fractionation/imaging | Predominantly nuclear [1] | Punctate cytoplasmic, excluded from nucleus [1] | [1] |
| Kinase Activation | Phosphorylation (Ser281/282, Thr444/442) | Activated by membrane-targeted hMOBs [8] | Activated by membrane-targeted hMOBs [8] | [8] |
| Neuronal Development | Retinal cell layers in knockout mice | Deletion leads to apoptosis and proliferation in retina [5] | Deletion increases mitochondrial oxidative stress genes [5] | [5] |
Detailed Experimental Protocols for NDR1/2 Research
Protocol 1: Investigating Subcellular Localization and Activation
Objective: To determine the differential localization and activation mechanisms of NDR1 and NDR2 kinases.
Methodology Details:
- Cell Culture and Transfection: Culture COS-7, HEK293, or HeLa cells in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal calf serum. Plate cells at consistent confluence (e.g., 3 × 10^5 cells/6-cm dish) and transfect the following day using Fugene 6 or Lipofectamine 2000 according to manufacturer instructions [8].
- Plasmid Construction: Generate N-terminal or C-terminal epitope-tagged (e.g., HA, myc) constructs of NDR1 and NDR2 in mammalian expression vectors. To assess localization importance, create chimeric constructs with altered targeting sequences (e.g., membrane-targeted using Lck tyrosine kinase motif, or nuclear-targeted using SV40 NLS) [8].
- Activation and Inhibition: To study activation, treat cells with 1 μM okadaic acid for 60 minutes to inhibit protein phosphatase 2A (PP2A), which regulates NDR phosphorylation status. For specific pathway activation, serum-starve cells overnight before stimulation with 100 ng/ml 12-O-tetradecanoylphorbol 13-acetate [8].
- Imaging and Biochemical Analysis: Fix cells and process for immunofluorescence microscopy using anti-tag and phospho-specific antibodies. For biochemical analysis, perform subcellular fractionation to separate nuclear and cytoplasmic components, followed by immunoblotting with antibodies against NDR1, NDR2, and their phosphorylated forms (e.g., anti-T444-P) [8].
Protocol 2: Assessing TLR9-Mediated Innate Immune Response
Objective: To evaluate the role of NDR1/2 in pattern recognition receptor signaling, specifically TLR9-mediated inflammation.
Methodology Details:
- Cell Model Preparation: Isolate primary macrophages from wild-type and Stk38-deficient mice. Alternatively, use immortalized macrophage cell lines with siRNA-mediated knockdown of NDR1 or NDR2 [3].
- Stimulation and Inhibition: Stimulate cells with CpG DNA (a TLR9 ligand) at optimized concentrations. For mechanistic studies, utilize pharmacological inhibitors targeting downstream signaling components (e.g., MEK/ERK pathway) [3].
- Interaction Studies: Perform co-immunoprecipitation experiments to investigate protein complexes. Transfect cells with tagged NDR1/2 constructs, immunoprecipitate with tag-specific antibodies, and probe for interacting proteins like the E3 ubiquitin ligase Smurf1 and its substrate MEKK2 [3].
- Output Measurements: Collect culture supernatants at various time points post-stimulation and measure cytokine production (TNF-α, IL-6) via ELISA. Analyze cell lysates by Western blot to detect phosphorylation of ERK1/2 and degradation of MEKK2 [3].
Signaling Pathways and Molecular Interactions
The functional differences between NDR1 and NDR2 are embedded within distinct signaling networks that regulate their activity and downstream effects. The following diagrams illustrate key pathways involving these kinases in neuroimmune contexts.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for NDR1/2 Research
| Reagent Category | Specific Examples | Function and Application |
|---|---|---|
| Cell Lines | HeLa, HEK293, COS-7, U2-OS, Primary Murine Macrophages/Microglia | Model systems for localization, interaction, and functional studies [3] [8] |
| Expression Constructs | Epitope-tagged NDR1/2 (HA, myc), Membrane-targeted (Lck motif), Nuclear-targeted (SV40 NLS), MOB isoforms | Investigate subcellular localization, activation mechanisms, and protein-protein interactions [8] |
| Activation/Inhibition Reagents | Okadaic acid (PP2A inhibitor), TPA, Leptomycin B (nuclear export inhibitor) | Modulate kinase phosphorylation status and study activation requirements [8] |
| Antibodies | Anti-NDR1, Anti-NDR2, Phospho-specific (Ser281, Thr444), Anti-HA, Anti-myc, Anti-MOBI/2 | Detection of protein expression, phosphorylation status, and localization via Western blot and immunofluorescence [8] |
| Cytokine/Chemokine Assays | ELISA for TNF-α, IL-6, Type I IFNs | Quantify inflammatory outputs in immune response studies [3] |
| Gene Manipulation Tools | siRNA for knockdown, CRISPR/Cas9 for knockout, Transgenic mouse models (Stk38-deficient) | Investigate loss-of-function phenotypes and validate physiological relevance [3] |
| ZQ-16 | ZQ-16|GPR84 Agonist |
The distinct subcellular localization of NDR1 (nuclear) and NDR2 (cytoplasmic) underpins their specialized functions in neuroimmune regulation, with NDR1 predominantly fine-tuning transcriptional responses and NDR2 modulating cytoplasmic signaling complexes. The optimal model system for NDR kinase research depends heavily on the specific biological question, ranging from reductionist cell lines for mechanistic studies to complex animal models for physiological validation. As the field advances, the development of more sophisticated 3D and microfluidic systems promises to bridge the gap between cellular mechanisms and tissue-level neuroimmune pathophysiology, potentially offering new insights into how the NDR1/2 axis could be targeted therapeutically in neurological and inflammatory disorders.
Resolving Key Challenges in NDR1/2 Research: Specificity, Localization, and Functional Overlap
The serine-threonine kinases NDR1 (STK38) and NDR2 (STK38L) share approximately 87% amino acid sequence identity, presenting a significant challenge for specific antibody-based detection [4] [6]. Despite this high similarity, these kinases exhibit distinct subcellular localizations and non-overlapping functions in key biological processes. NDR1 demonstrates diffuse nuclear and cytoplasmic distribution, while NDR2 exhibits punctate cytoplasmic localization, primarily targeting peroxisomes [4] [6]. This technical comparison guide objectively evaluates antibody performance for discriminating between these highly similar kinases, providing researchers with validated experimental approaches to overcome cross-reactivity pitfalls.
Comparative Localization and Functional Differences
Table 1: Fundamental Differences Between NDR1 and NDR2
| Characteristic | NDR1 (STK38) | NDR2 (STK38L) |
|---|---|---|
| Subcellular Localization | Diffuse nuclear and cytoplasmic [4] [6] | Punctate cytoplasmic (peroxisomal) [4] |
| C-terminal Sequence | Ala-Lys [4] | Gly-Lys-Leu (GKL; PTS1-like) [4] |
| Pex5p Binding | No binding [4] | Direct binding [4] |
| Role in Ciliogenesis | Not involved [4] | Essential (phosphorylates Rabin8) [4] |
| Expression Profile | Widely expressed [6] | Highest in thymus [6] |
Documented Cross-Reactivity Pitfalls and Evidence
Challenges with Commercial Antibodies
Multiple studies have demonstrated that improperly validated antibodies contribute to erroneous conclusions about NDR1/NDR2 localization and function:
Non-specific N-terminal antibodies: Several commercial antibodies targeting the N-terminal region (aa 1-100) fail to distinguish between NDR1 and NDR2 due to high sequence conservation in this region [12]. For example, antibody E-2 (#sc-271703) detects both kinases in human microglial cells [12].
C-terminal specificity advantages: Antibodies generated against the C-terminal regions show improved specificity. One study successfully used an NDR2-specific antibody targeting amino acids 380-460 (#STJ94368), which did not cross-react with NDR1 [12].
Validation inadequacies: Many commercially available antibodies lack sufficient validation using knockout controls, leading to misinterpretation of immunohistochemistry and Western blot results [38].
Consequences of Misinterpretation
The functional implications of antibody cross-reactivity are significant. Before NDR2's peroxisomal localization was established, its punctate pattern was sometimes misinterpreted as endosomal or Golgi localization [4]. Similarly, studies of NDR1's nuclear function may be confounded by NDR2 detection if cross-reactive antibodies are used [6].
Recommended Validation Methodologies
Knockout Validation Protocol
Purpose: To confirm antibody specificity using genetic controls. Materials: Wild-type and NDR1/NDR2 knockout cell lines or tissues. Procedure:
- Generate or obtain NDR1 and NDR2 knockout models [38]
- Prepare protein lysates from wild-type and knockout cells
- Perform Western blotting using candidate antibodies
- Compare signal intensity between samples Interpretation: A specific NDR1 antibody should show diminished signal only in NDR1 KO samples, not in NDR2 KO samples [38].
Subcellular Fractionation with Validation
Purpose: To distinguish true nuclear NDR1 from cytoplasmic NDR2. Materials: Subcellular fractionation kit, protease inhibitors, centrifugation equipment. Procedure:
- Fractionate cells into nuclear, cytoplasmic, and organellar fractions [4]
- Validate fraction purity using marker proteins (e.g., Lamin A/C for nuclear fraction)
- Perform Western blotting with candidate antibodies
- Compare fractionation patterns with known localization [4] Expected Results: NDR1 should enrich in nuclear fractions, while NDR2 should enrich in peroxisome-containing fractions [4].
Immunofluorescence Co-localization Protocol
Purpose: To verify subcellular localization using multiple markers. Materials: Fixed cells, validated primary antibodies, species-specific fluorescent secondary antibodies, confocal microscope. Procedure:
- Culture cells on glass coverslips
- Fix with 4% paraformaldehyde and permeabilize with 0.1% Triton X-100
- Incubate with primary antibodies against NDR1/NDR2 and organelle markers (e.g., catalase for peroxisomes)
- Incubate with fluorescent secondary antibodies
- Image using confocal microscopy and quantify co-localization [4] Validation: NDR2 should show >80% co-localization with peroxisomal markers like CFP-SKL or catalase [4].
Experimental Data on Antibody Performance
Table 2: Antibody Validation Data and Performance Metrics
| Antibody Target | Source/Reference | Specificity Confirmed | Applications | Key Findings |
|---|---|---|---|---|
| NDR1/2 (N-terminal) | #sc-271703 [12] | No (cross-reactive) | ICC (human) | Detects both NDR1 and NDR2 in microglia |
| NDR2 (C-terminal) | #STJ94368 [12] | Yes | ICC, WB | Specific for NDR2 in mouse/human microglia |
| NDR1-specific | In-house [2] | Yes | WB, ICC | No detection of overexpressed NDR2 |
| NDR2-specific | In-house [2] | Yes | WB, ICC | No detection of overexpressed NDR1 |
| Anti-NDR CT | [8] | Limited | IP, WB | May cross-react depending on immunogen |
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for NDR1/NDR2 Research
| Reagent | Function/Application | Example Sources/References |
|---|---|---|
| CFP-SKL | Peroxisome marker for co-localization | [4] |
| Catalase antibody | Peroxisome validation | [4] |
| Pex5p reagents | NDR2 interaction studies | [4] |
| MOB1/2 proteins | NDR kinase activators | [6] [8] |
| CRISPR-Cas9 kits | Generation of knockout validation models | [12] [38] |
| Subcellular fractionation kits | Localization validation | [4] |
| Okadaic acid | NDR kinase activator (PP2A inhibitor) | [8] |
Decision Framework for Antibody Selection
Antibody Selection Workflow
The high sequence similarity between NDR1 and NDR2 demands rigorous antibody validation to prevent erroneous conclusions in subcellular localization and functional studies. Researchers should prioritize C-terminal specific antibodies when available and implement knockout validation, subcellular fractionation, and co-localization studies to confirm specificity. Proper antibody validation is not merely a technical consideration but a fundamental requirement for accurate interpretation of NDR kinase biology, particularly given their distinct roles in processes ranging from ciliogenesis to cancer progression. The methodologies and frameworks presented here provide researchers with tools to navigate these challenges effectively.
The canonical view of NDR kinase localization, which posits a nuclear distribution for NDR1 and a cytoplasmic distribution for NDR2, provides a foundational framework for understanding their distinct cellular functions [14]. This spatial segregation suggests divergent roles in cellular signaling and has significant implications for research methodologies and interpretation. However, a growing body of evidence indicates that this binary localization model represents an oversimplification. The subcellular distribution of NDR kinases demonstrates remarkable plasticity, influenced by cell type-specific factors, developmental stages, and particularly by various cellular stress conditions. This article systematically examines the discrepancies in NDR localization studies, highlighting how experimental context influences observed distribution patterns and providing methodological guidance for accurate interpretation of localization data in physiological and pathological contexts, including cancer and inflammatory diseases.
Comparative Analysis of NDR1 and NDR2 Localization Across Experimental Conditions
Table 1: Summary of NDR1 and NDR2 Localization Across Cell Types and Conditions
| Kinase | Canonical Localization | Condition/Cell Type | Observed Localization Shift | Functional Consequences | Citation |
|---|---|---|---|---|---|
| NDR1 | Nuclear | General Localization | Nucleus | Regulation of cell cycle progression, apoptosis | [14] |
| NDR2 | Cytoplasmic | General Localization | Cytoplasm | Regulation of cytoskeletal dynamics, vesicle trafficking | [14] |
| NDR2 | Cytoplasmic | BV-2 Microglial Cells (Normal Glucose) | Cytoplasmic, peri-nuclear pattern | Baseline cellular homeostasis | [12] |
| NDR2 | Cytoplasmic | BV-2 Microglial Cells (High Glucose) | Cell periphery and tips of microglial processes | Enhanced migratory and damage-sensing capacity | [12] |
| NDR2 | Cytoplasmic | Mouse Primary Retinal Microglia | Predominantly cell periphery and tips of processes | Potential role in immunological surveillance | [12] |
| NDR1/NDR2 | Nuclear/Cytoplasmic | Human iPSC-Derived Microglial Cells | Colocalization with cytoskeletal protein IBA1 | Association with cytoskeletal remodeling machinery | [12] |
The data presented in Table 1 reveals several critical patterns. First, the canonical nuclear/cytoplasmic dichotomy between NDR1 and NDR2 is consistently observed in standard cell culture models. However, specialized cell types, particularly those of neural origin like microglia, exhibit markedly different localization patterns. Furthermore, metabolic stress conditions, such as high glucose exposure, can dynamically redistribute NDR kinases to specific subcellular compartments, potentially altering their functional roles in adaptive responses.
Impact of Stress Conditions on NDR Localization and Function
Metabolic Stress: High Glucose Conditions
Under high glucose conditions mimicking diabetic stress, NDR2 protein expression is significantly upregulated in BV-2 microglial cells, with a notable redistribution to the cell periphery and tips of microglial processes [12]. This redistribution coincides with functional impairments in mitochondrial respiration, metabolic flexibility, phagocytic capacity, and migratory ability. Interestingly, this localization shift occurs without immediate alterations in Ndr2 mRNA levels, suggesting post-translational regulation mechanisms [12]. The spatial repositioning of NDR2 under metabolic stress implies its involvement in adapting cellular functions to pathological conditions, particularly in the context of diabetic retinopathy.
Inflammatory and Infectious Stress
NDR kinases demonstrate complex localization and functional adaptations during inflammatory and infectious challenges. NDR1 functions as a negative regulator of TLR9-mediated immune response in macrophages by promoting ubiquitination and degradation of MEKK2, thereby inhibiting CpG-induced ERK1/2 activation and subsequent pro-inflammatory cytokine production [14]. Conversely, in antiviral immunity, NDR1 enhances type I interferon production by binding to the intergenic region of miR146a and promoting STAT1 translation [14]. Simultaneously, NDR2 promotes RIG-I-mediated antiviral response by facilitating the formation of the RIG-I/TRIM25 complex and enhancing K63-linked polyubiquitination of RIG-I [14]. These differential roles in distinct immune pathways suggest potential context-dependent subcellular redistribution that requires further investigation.
Methodological Considerations for Localization Studies
Key Experimental Protocols
Table 2: Essential Research Reagents and Methodologies for NDR Localization Studies
| Research Reagent/Method | Specific Application | Technical Considerations | Function in Experimental Design |
|---|---|---|---|
| Immunocytochemistry | Protein localization visualization | Antibody specificity validation critical; fixation methods affect epitope availability | Spatial distribution analysis in fixed cells |
| Commercial NDR1/2 Antibody (E-2) #sc-271703 | Targeting N-terminus (aa 1-100) of human NDR2 | Validated for human iPSC-derived microglial cultures | Detection of native NDR kinase expression |
| Commercial NDR2 Antibody #STJ94368 | Targeting C-terminus (aa 380-460) of human NDR2 | Effective in mouse primary and immortalized microglial cells | Species-specific application considerations |
| CRISPR-Cas9 Gene Editing | NDR kinase knockdown/knockout | Partial knockdown essential for viability in some cell types | Functional validation of localization observations |
| Western Blot Analysis | Protein expression quantification | Normalization to calnexin recommended; multiple exposure times needed | Quantitative assessment of expression changes |
| siRNA-mediated knockdown | Transient reduction of NDR expression | Double transfection at 24-h intervals may enhance efficiency | Rapid assessment of kinase function |
| qRT-PCR | mRNA expression analysis | No immediate Ndr2 mRNA changes under high glucose despite protein increases | Discrimination between transcriptional and translational regulation |
Detailed Experimental Workflow for Comprehensive Localization Studies
Cell Culture and Treatment Conditions:
- Maintain relevant cell lines (e.g., BV-2 microglial cells, iPSC-derived microglia, primary retinal microglia) in appropriate media supplemented with 10% FCS [27].
- Apply stress conditions (e.g., 30.5 mM glucose for high glucose exposure) for defined periods (7h to 12h) [12].
- Include proper controls (normal glucose, 5.5 mM) for comparison.
Protein Localization Analysis:
- Perform immunocytochemistry using validated antibodies targeting specific NDR epitopes [12].
- Co-stain with cell-type specific markers (e.g., IBA1 for microglia, NeuN for neurons, GFAP for astrocytes) to verify cell identity and assess co-localization.
- Utilize confocal microscopy for high-resolution subcellular localization.
Functional Validation:
- Implement CRISPR-Cas9 or siRNA-mediated knockdown to perturb NDR expression [12].
- Assess functional consequences through migration assays, phagocytosis assays, metabolic measurements, and cytokine profiling.
- Correlate localization changes with functional outcomes.
Expression Analysis:
- Conduct Western blotting to quantify protein expression changes under different conditions.
- Perform qRT-PCR to discriminate between transcriptional and translational regulation.
- Use cycloheximide chase experiments to assess protein stability when appropriate.
Diagram Title: Comprehensive Workflow for NDR Localization Studies
Signaling Pathways Influenced by NDR Localization
Diagram Title: NDR Kinase Signaling Networks and Functional Roles
The investigation of NDR kinase localization reveals a complex regulatory landscape where cell type, stress conditions, and methodological approaches significantly influence experimental outcomes. The canonical nuclear/cytoplasmic partitioning of NDR1/NDR2 represents a simplified model that requires contextual refinement. Researchers should adopt the following practices to minimize interpretive errors:
- Validate findings across multiple cell types, including primary cells relevant to the physiological or pathological context.
- Include stress conditions appropriate to the research question, particularly metabolic and inflammatory challenges.
- Employ multiple complementary techniques for localization studies, including immunocytochemistry with validated antibodies, biochemical fractionation, and live-cell imaging where possible.
- Correlate localization patterns with functional assays to establish physiological relevance.
- Consider species-specific differences in NDR kinase expression and localization when translating findings across model systems.
Understanding the dynamic nature of NDR kinase localization and its functional implications will enhance our comprehension of their roles in health and disease, particularly in conditions like cancer, diabetic retinopathy, and inflammatory disorders where these kinases emerge as potential therapeutic targets.
NDR kinase research provides a paradigm for understanding a critical challenge in genetic research: compensatory mechanisms between homologous genes can completely mask the true biological function of a gene in single-knockout (KO) models. While single KO of either Ndr1 or Ndr2 in mice yields minimal phenotypic consequences, double KO studies reveal these kinases are essential for embryonic development, regulating processes including somitogenesis and cardiac looping [50]. This guide objectively compares experimental approaches for characterizing such compensatory mechanisms, using the NDR kinase family as a primary case study to provide methodological frameworks applicable to genetic research across biological systems.
Comparative Phenotypic Analysis: Single vs. Double Knockout Models
The stark contrast between single and double knockout phenotypes provides the most direct evidence for functional compensation.
Table 1: Phenotypic Consequences of NDR Kinase Inactivation in Mouse Models
| Genetic Model | Viability & Development | Specific Tissue Phenotypes | Molecular Compensation Evidence |
|---|---|---|---|
| Ndr1 Single KO | Viable, fertile, normal lifespan [50] | Predisposition to T-cell lymphoma [51] | NDR2 protein upregulated in Ndr1 KO tissues [51] [50] |
| Ndr2 Single KO | Viable, fertile, normal lifespan [50] | Hyperplastic growth in intestinal epithelium; increased susceptibility to colon carcinogenesis [51] | NDR1 protein levels increased in colon tissue [50] |
| Ndr1/2 Double KO | Embryonic lethality by ~E10.5 [50] | Severe defects in somite patterning, arrested cardiac looping, pericardial edema [50] | Compensation not possible; reveals essential developmental functions |
Experimental Strategies for Unmasking Compensation
Genetic Workflow for Comprehensive KO Analysis
The following diagram outlines the sequential genetic strategy to test for compensatory mechanisms between homologous genes.
Methodological Framework for Detecting Compensation
Protein-Level Compensation Analysis
Objective: Determine whether loss of one gene isoform leads to upregulated expression of its homolog.
Protocol:
- Generate KO Models: Create single KO models for each gene homolog (Ndr1 KO and Ndr2 KO) and a double KO model using Cre-loxP technology [51] [50].
- Tissue Collection: Harvest multiple tissues representing different expression patterns (e.g., thymus for high NDR1, colon for high NDR2).
- Protein Extraction and Western Blotting:
- Quantification: Normalize protein levels to loading controls; compare expression levels between wild-type, single KO, and double KO tissues.
Expected Outcome: In Ndr1 KO tissues, NDR2 protein levels increase; conversely, Ndr2 KO shows elevated NDR1 in specific tissues [51] [50].
Functional Phenotypic Analysis in Double Mutants
Objective: Reveal essential biological functions masked by compensation in single KOs.
Protocol:
- Embryonic Timed Matings: Set up heterozygous double mutant crosses to obtain double null embryos.
- Genotype Analysis: Use PCR-based genotyping of embryonic tissues [50].
- Histological Analysis:
- Process embryos for histological sectioning and H&E staining.
- Examine specific developmental processes: somite formation, cardiac looping, neural tube closure.
- Molecular Marker Analysis:
- Perform whole-mount in situ hybridization for key pathway genes (e.g., Lunatic fringe for Notch signaling in somitogenesis) [50].
- Analyze expression patterns for symmetry and intensity compared to wild-type.
Expected Outcome: Ndr1/2 double null embryos reveal essential roles in somitogenesis and cardiac looping, demonstrating the developmental processes requiring NDR kinase function [50].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Genetic Compensation Studies
| Reagent/Cell Line | Specific Function | Application in NDR Studies |
|---|---|---|
| Isoform-Specific Antibodies | Distinguish between highly similar protein homologs (87% identity for NDR1/2) | Detect protein-level compensation; NDR2 upregulation in Ndr1 KO tissues [51] |
| Conditional KO Models (Cre-loxP) | Enable tissue-specific and time-controlled gene inactivation | Study gene function in specific tissues (e.g., intestinal epithelium) without embryonic lethality [51] |
| Phospho-Specific Antibodies | Assess kinase activation state (e.g., hydrophobic motif phosphorylation) | Monitor functional activity of compensating kinases [50] |
| CRISPR/Cas9 Systems | Generate complete gene knockouts with high efficiency | Create single and double KO cell lines for in vitro validation [52] |
| Primary Cell Isolation | Study cell behavior ex vivo under defined conditions | Assess proliferation changes in KO intestinal epithelial cells [51] |
NDR-Specific Compensatory Mechanisms and Technical Considerations
Distinct Functions Despite High Similarity
Although NDR1 and NDR2 share 87% amino acid identity and overlapping functions in many contexts, they also perform distinct physiological roles due to differences in:
- Subcellular localization: NDR2 localizes to peroxisomes via a C-terminal Gly-Lys-Leu motif, while NDR1 displays diffuse cytoplasmic distribution [4].
- Tissue-specific expression: NDR1 predominates in immune tissues, while NDR2 shows highest expression in gastrointestinal tract and brain [51] [4].
- Specialized functions: NDR2, but not NDR1, is essential for primary cilium formation through Rabin8 phosphorylation and regulation of vesicular trafficking [4].
Common Artifacts and Mitigation Strategies
- Incomplete KO Models: Gene-trap alleles may produce functional truncated proteins. Use multiple independent KO strategies to confirm phenotypes [50].
- Strain-Specific Effects: Genetic background can influence compensatory responses. Conduct studies on congenic backgrounds.
- Temporal Considerations: Compensation may occur at different developmental stages. Implement inducible KO systems to bypass developmental compensation.
The NDR kinase model demonstrates that functional compensation between homologous genes represents a fundamental biological phenomenon that can completely obscure gene function in conventional single-KO studies. Researchers investigating gene families with high sequence similarity should implement systematic double-KO approaches coupled with protein-level expression analysis to uncover authentic biological functions. The methodological framework presented here provides a standardized approach for distinguishing between genuine genetic redundancy and technical artifacts in functional genetic studies, with particular relevance for drug development targeting signaling pathways with multiple homologous components.
In the study of NDR kinases, a central challenge is unequivocally identifying their direct phosphorylation targets amidst a cascade of downstream, indirect effects. The high sequence similarity between NDR1 and NDR2, coupled with their distinct subcellular localizations—nuclear versus cytoplasmic/peroxisomal—further complicates this task, as identical kinase domains may encounter different substrate pools. This guide outlines strategies to overcome this pitfall, using comparative studies of NDR1 and NDR2 as a framework.
Comparative Profile: NDR1 vs. NDR2
The functional divergence between NDR1 and NDR2 is heavily influenced by their distinct subcellular localizations, which dictates substrate accessibility. The table below summarizes key differentiating features.
| Feature | NDR1 | NDR2 |
|---|---|---|
| Primary Subcellular Localization | Nucleus, Cytoplasm [8] | Cytoplasm, Peroxisomes [4], Cell periphery/process tips (microglia) [12] |
| Key Localization Signal | Nuclear Localization Signal (NLS) [8] | Peroxisomal Targeting Signal 1 (PTS1), C-terminal Gly-Lys-Leu (GKL) [4] |
| Impact of Localization on Function | Suggests role in nuclear processes [8] | Essential for ciliogenesis via Rabin8 phosphorylation at peroxisomes; regulates microglial metabolism and inflammation [12] [4] |
| Co-activator Binding | Binds hMOB proteins; activation at plasma membrane [8] [9] | Binds hMOB proteins; activation at plasma membrane [8] |
Experimental Protocol to Disentangle Direct Effects
Mass spectrometry-based phosphoproteomics is the cornerstone for identifying kinase substrates. The following workflow, adapted from studies on protein phosphatase-1 (PP1), provides a robust method to differentiate direct NDR substrates from indirect phosphorylation events [53].
Protocol Details:
- Cellular Treatment & Rapid Analysis: Stimulate cells in a time-controlled manner to activate NDR kinases. This can be achieved by:
- Pharmacological inhibition of counteracting phosphatases with Okadaic Acid (OA) [8].
- Inducible translocation of activating co-factors like hMOB to the membrane [8].
- Crucially, use very short stimulation times (e.g., minutes) after activator introduction. This focuses the analysis on the earliest phosphorylation events, minimizing the accumulation of secondary, indirect effects [53].
- Phosphoproteomics (LC-MS/MS): Lyse cells and analyze protein phosphorylation changes using liquid chromatography with tandem mass spectrometry (LC-MS/MS). Compare treated samples to controls to identify sites that show significant changes [53].
- In Vitro Kinase Assay: To confirm direct phosphorylation, take the candidate substrates identified in step 2 and incubate them with purified NDR kinase in a test tube. A positive result here confirms a direct kinase-substrate relationship, as no other cellular components are present [53].
- Data Integration: Overlap the results from the cellular and in vitro experiments. Phosphosites that are rapidly induced in cells and can be phosphorylated by NDR in vitro represent high-confidence direct substrates.
The Scientist's Toolkit: Essential Research Reagents
The following reagents are critical for studying NDR kinase function and substrate identification.
| Research Reagent / Tool | Function / Application | Key Detail / Target |
|---|---|---|
| Okadaic Acid (OA) | Activates NDR kinases by inhibiting Protein Phosphatase 2A (PP2A), leading to increased phosphorylation of Thr444/Thr442 and Ser281/Ser282 [8]. | NDR1/2 Activator |
| Membrane-Targeted hMOB | Constitutively activates NDR by recruiting it to the plasma membrane, used to study MOB-dependent activation mechanisms [8]. | NDR1/2 Co-activator |
| PDPs (PP1-Disrupting Peptides) | Chemical probes that release active PP1 catalytic subunits from specific holoenzymes; used as a model for studying direct vs. indirect dephosphorylation events [53]. | PP1 Activator / Probe |
| Anti-NDR CT Antibody | Detects total NDR protein levels [8]. | NDR1/2 Detection |
| Anti-phospho-NDR Antibodies | Specifically recognizes activated NDR1 (phospho-Thr444, phospho-Ser281); critical for assessing kinase activity state [8]. | Active NDR1 Detection |
| CRISPR-Cas9 (Ndr2/Stk38l sgRNA) | Generates genetically modified cell lines (e.g., in BV-2 microglia) to study NDR2-specific loss-of-function phenotypes [12]. | NDR2 Gene Knockout |
| Pex5p Knockdown | Inhibits peroxisomal protein import; used to dissect the dependency of NDR2 functions on its peroxisomal localization [4]. | Peroxisome Function Disruption |
Visualizing NDR Activation and Localization
The distinct activation mechanisms and localization of NDR1 and NDR2 are key to understanding their functional differences. The pathway below integrates these concepts.
By applying these targeted experimental strategies, researchers can more effectively pinpoint the direct substrates of NDR1 and NDR2, moving beyond correlative phosphorylation events to establish causal relationships. This precision is fundamental to understanding how their spatial regulation translates into distinct physiological and pathological roles.
The functional characterization of proteins within cellular pathways presents a significant challenge in molecular biology. This is particularly true for highly homologous proteins like NDR1 (STK38) and NDR2 (STK38L), serine-threonine kinases sharing approximately 87% amino acid sequence identity yet demonstrating distinct subcellular localizations and non-overlapping functions in key cellular processes [4] [6] [10]. Despite their close relationship, NDR1 exhibits diffuse nuclear and cytoplasmic distribution, while NDR2 shows a punctate cytoplasmic pattern [4] [6]. This guide objectively compares experimental strategies used to dissect these functional differences, providing a framework for researchers investigating paralogous proteins. We focus on the integration of genetic, biochemical, and cell biological approaches, using the resolved question of NDR2's peroxisomal localization and its implications for ciliogenesis as a central case study [4].
Comparative Analysis of NDR1 and NDR2 Localization and Function
Table 1: Key Characteristics of NDR1 and NDR2 Kinases
| Feature | NDR1 (STK38) | NDR2 (STK38L) |
|---|---|---|
| Amino Acid Sequence Identity | ~87% to NDR2 [4] [10] | ~87% to NDR1 [4] [10] |
| Subcellular Localization | Diffuse nuclear and cytoplasmic distribution [4] [6] | Punctate cytoplasmic distribution; localizes to peroxisomes [4] |
| C-Terminal Sequence | Ends in Ala-Lys [4] | Ends in Gly-Lys-Leu (GKL) - a PTS1-like sequence [4] |
| Interaction with Pex5p | Does not bind [4] | Binds to the PTS1 receptor [4] |
| Role in Ciliogenesis | No significant role; depletion does not suppress it [4] | Critical role; depletion significantly suppresses it [4] |
| Expression in Microglia | Detected in human iPSC-derived microglia [12] | Expressed in microglia; upregulated under high-glucose conditions [12] |
| Oncogenic/Tumor Suppressor Role | Predisposition to T-cell lymphoma in knock-out mice [4] | Behaves as an oncogene in most cancers (e.g., lung cancer) [10] |
Table 2: Functional Consequences of NDR2 Perturbation in Different Cell Types
| Cell Type | Experimental Perturbation | Key Observed Phenotypes | Supporting Data |
|---|---|---|---|
| Retinal Pigment Epithelial (RPE1) Cells | Knockdown of NDR2 [4] | Suppressed primary cilium formation [4] | Immunofluorescence, rescue experiments |
| BV-2 Microglial Cells | Partial CRISPR-Cas9 KO under high glucose [12] | Impaired mitochondrial respiration, reduced phagocytosis and migration, elevated pro-inflammatory cytokines [12] | Seahorse analysis, phagocytosis/migration assays, ELISA/qPCR |
| Mouse Retinal Cells | Single knockout (Ndr2(^{-/-})) [38] | Aberrant rod opsin localization, increased cell proliferation in INL, decreased amacrine cells, reduced Aak1 levels [38] | Immunohistochemistry, transcriptome analysis, immunoblotting |
| Canine Retinal Cells | Naturally occurring Ndr2 mutation (erd) [38] | Early retinal degeneration, photoreceptor proliferation & apoptosis, retinal disorganization [38] | Histopathology, genetic analysis |
Experimental Protocols for Differentiating NDR1/NDR2 Function
Protocol 1: Determining Subcellular Localization via Co-Immunostaining
This protocol is foundational for establishing the distinct cytoplasmic distributions of NDR1 and NDR2 [4].
- 1. Cell Seeding and Transfection: Seed appropriate cells (e.g., RPE1 or HeLa) on glass coverslips. Transfect with plasmids encoding fluorescently tagged proteins (e.g., YFP-NDR2 and CFP-SKL) [4].
- 2. Fixation and Permeabilization: After 24-48 hours, fix cells with 4% paraformaldehyde for 15 minutes at room temperature. Permeabilize with 0.1% Triton X-100 in PBS for 10 minutes [4].
- 3. Immunostaining: Incubate cells with primary antibodies against organelle markers (e.g., Catalase for peroxisomes, EEA1 for early endosomes, GM130 for Golgi) for 1 hour. After washing, incubate with fluorophore-conjugated secondary antibodies for 45 minutes [4].
- 4. Imaging and Analysis: Mount coverslips and image using a confocal microscope. Analyze co-localization using software such as ImageJ with appropriate plugins (e.g., JaCoP) to calculate Pearson's correlation coefficient or Mander's overlap coefficient [4].
Protocol 2: Functional Validation of a Targeting Sequence
This biochemical and genetic protocol confirms the role of a specific motif, such as the C-terminal GKL of NDR2 [4].
- 1. Mutagenesis: Generate a mutant construct of NDR2 where the C-terminal leucine is deleted (NDR2(ΔL)) using site-directed mutagenesis [4].
- 2. Localization Analysis: Transfect the wild-type (YFP-NDR2) and mutant (YFP-NDR2(ΔL)) constructs into cells and perform immunostaining as in Protocol 1. The diffuse distribution of the mutant confirms the necessity of the complete tripeptide for punctate/peroxisomal targeting [4].
- 3. Interaction Assay:
- Cell Lysis: Lyse cells expressing YFP-NDR2, YFP-NDR1, or YFP-NDR2(ΔL) in a non-denaturing lysis buffer.
- Co-Immunoprecipitation (Co-IP): Use an anti-GFP antibody to immunoprecipitate the tagged proteins and their associated complexes from the cell lysates.
- Immunoblotting: Probe the immunoprecipitates with an antibody against the PTS1 receptor, Pex5p. Binding of Pex5p to NDR2, but not to NDR1 or NDR2(ΔL), validates the specific molecular interaction [4].
- 4. Functional Rescue: In cells where endogenous NDR2 has been knocked down, re-express either wild-type NDR2 or the NDR2(ΔL) mutant. Assess the rescue of the functional phenotype (e.g., ciliogenesis efficiency) to link the targeting motif to biological function [4].
Protocol 3: Genetic Optimization for Pathway Analysis
Genetic algorithms (GAs) provide a powerful, knowledge-independent method for optimizing complex biological systems, applicable to media optimization or genetic interaction studies [54] [55].
- 1. Initialization: Define the "genes" (e.g., concentrations of amino acids, expression levels of pathway genes) and create an initial population of "individuals" (unique combinations of these parameters) [54] [55].
- 2. Fitness Evaluation: Test each individual in the biological system (e.g., cell culture) and measure its "fitness" (e.g., metabolic activity, metabolite production) [54].
- 3. Selection: Select the top-performing individuals to be "parents" for the next generation. An elitist strategy can be used to carry the best performer forward directly [55].
- 4. Crossover and Mutation: Create a new generation of individuals by combining parts of the "chromosomes" of two parents (crossover) and by randomly altering bits within a chromosome (mutation) [54] [55].
- 5. Iteration: Repeat steps 2-4 for multiple generations. The population evolves toward an optimal combination of parameters, which can then be validated and mechanistically studied [54].
Integrated Signaling Pathways and Functional Workflows
Understanding how distinct subcellular localization drives functional specificity requires mapping the pathways involved.
The Scientist's Toolkit: Key Research Reagents and Solutions
Table 3: Essential Reagents for Investigating NDR1/NDR2 Localization and Function
| Reagent / Solution | Function / Application | Example / Note |
|---|---|---|
| Plasmids: Fluorescently Tagged NDR1/NDR2 | Visualizing subcellular localization and dynamics in live or fixed cells. | YFP-NDR2, CFP-NDR1; NDR2(ΔL) mutant for motif validation [4]. |
| Pex5p Antibody | Detecting the PTS1 receptor; used in Co-IP to confirm interaction with NDR2. | Critical for validating the peroxisomal targeting mechanism [4]. |
| Organelle Marker Antibodies | Identifying specific organelles in co-localization studies. | Catalase (peroxisomes), EEA1 (endosomes), GM130 (Golgi) [4]. |
| CRISPR-Cas9 System | Generating stable knockout or knockdown cell lines for functional studies. | Used in BV-2 microglia to study NDR2 in metabolic adaptation [12]. |
| Mob2 Expression Construct | Activating NDR1/NDR2 kinases; essential for in vitro kinase assays. | Human Mob2 binds and dramatically stimulates NDR kinase activity [6]. |
| Artificial Sea Water (ASW) & CMF | Maintaining osmotic balance and dissociating cells in marine sponge studies. | Used in sponge cell culture medium optimization via genetic algorithms [54]. |
| Amino Acid Stock Solutions | Component for custom medium formulation in optimization strategies. | Prepared at 5 g/L for genetic algorithm-based medium optimization [54]. |
The distinct functions of NDR1 and NDR2, despite their high sequence similarity, underscore the critical importance of subcellular localization as a determinant of kinase specificity. Robust conclusions in this field are achieved not by a single approach, but by the strategic integration of multiple methods. Cell biology (localization studies), biochemistry (interaction assays), and genetics (CRISPR and genetic algorithms) form a powerful triad. The case of NDR2's peroxisomal targeting via its C-terminal GKL sequence and its subsequent role in ciliogenesis through Rabin8 phosphorylation exemplifies how this integrated strategy can resolve complex biological questions [4]. For researchers, this multi-faceted approach provides a validated blueprint for unraveling the functional nuances of other paralogous protein families, with significant implications for understanding cellular homeostasis and developing targeted therapeutic strategies.
Direct Functional Comparison: How NDR1 and NDR2 Orchestrate Distinct Biological Outcomes
The nuclear Dbf2-related (NDR) kinases NDR1 (STK38) and NDR2 (STK38L) are serine/threonine kinases belonging to the NDR/LATS subfamily of the Hippo signaling pathway [14] [3]. Despite their high sequence identity (approximately 87%), they exhibit distinct subcellular localizations that dictate their specialized functions in innate immunity [4] [6]. NDR1 is predominantly nuclear, whereas NDR2 is cytoplasmic and localizes to specific structures like peroxisomes [4] [6]. This fundamental difference in localization underpins their unique mechanisms of action: NDR1 functions as a transcriptional regulator in the nucleus to modulate immune responses, while NDR2 operates in the cytoplasm to directly regulate the activity of key signaling complexes, such as the RIG-I/TRIM25 complex [14] [56]. This guide provides a detailed, evidence-based comparison of their distinct roles, experimental methodologies, and functional outcomes in antiviral innate immunity.
Functional Comparison: Mechanisms and Experimental Evidence
Table 1: Functional Comparison of NDR1 and NDR2 in Innate Immunity
| Feature | NDR1 (STK38) | NDR2 (STK38L) |
|---|---|---|
| Subcellular Localization | Nucleus [14] [6] | Cytoplasm; punctate distribution associated with peroxisomes [4] [6] |
| Key Role in Antiviral Immunity | Transcriptional regulator of miR146a and STAT1 translator [14]; Positive regulator of type I and II IFN pathways [14] | Facilitates RIG-I/TRIM25 complex formation and K63-linked ubiquitination of RIG-I [14] [56] |
| Direct Binding Partners | Smurf1 [14], miR146a intergenic region (DNA) [14] | RIG-I, TRIM25 [56] |
| Downstream Signaling Target | MEKK2 (for TLR9 signaling) [14], STAT1 [14] | RIG-I [56] |
| Effect on Cytokine Production | Inhibits CpG-DNA (TLR9)-induced TNF-α and IL-6 [14]; Enhances virus-induced type I IFN and pro-inflammatory cytokines [14] | Promotes RNA virus-induced type I IFNs and pro-inflammatory cytokines [56] |
| In Vivo Phenotype (Knockout) | Stk38-deficient mice more susceptible to E. coli infection and polymicrobial sepsis; higher pro-inflammatory cytokine levels and mortality [14] | Conditional knockout mice (LysmâºNDR2f/f) show impaired antiviral immune response [56] |
NDR1: The Nuclear Transcriptional Regulator
NDR1 modulates the immune response through gene regulation within the nucleus. Its primary mechanism involves binding to the intergenic region of the microRNA miR146a, a key negative regulator of innate signaling. This binding dampens miR146a transcription, which in turn promotes the translation of the transcription factor STAT1. The increased STAT1 levels enhance the production of type I interferon (IFN), pro-inflammatory cytokines, and interferon-stimulated genes (ISGs), thereby bolstering the antiviral state [14]. This function of NDR1 occurs independently of its kinase activity. In the context of bacterial infection and TLR9 signaling, NDR1 acts as a negative regulator by forming a complex with the E3 ubiquitin ligase Smurf1 to promote the degradation of MEKK2, a kinase essential for ERK1/2 activation, thereby limiting pro-inflammatory cytokine production [14].
NDR2: The Cytoplasmic Complex Facilitator
NDR2 exerts its pro-antiviral function in the cytoplasm by directly interacting with the core components of the RIG-I signaling pathway. It associates with both the viral RNA sensor RIG-I and the E3 ubiquitin ligase TRIM25. This interaction facilitates the formation of the RIG-I/TRIM25 complex, which is crucial for the transfer of K63-linked polyubiquitin chains to RIG-I [56]. This specific ubiquitination is a critical switch that activates RIG-I, enabling it to oligomerize and interact with the downstream adapter MAVS on mitochondria, thereby initiating a signaling cascade that leads to robust type I IFN production [57] [58] [59]. The essential role of NDR2 in this pathway is confirmed by the impaired antiviral response observed in macrophage-specific Ndr2 knockout mice [56].
Experimental Protocols for Key Studies
Protocol: Investigating NDR1's Transcriptional Role
- Key Study: NDR1 binds to the intergenic region of miR146a and regulates STAT1 translation [14].
- Objective: To determine how NDR1 regulates the type I IFN pathway independently of its kinase activity.
- Methodology:
- Chromatin Immunoprecipitation (ChIP): Crosslink proteins to DNA in relevant cell lines (e.g., macrophages). Immunoprecipitate chromatin using an anti-NDR1 antibody. Purify the co-precipitated DNA and analyze the enrichment of the miR146a intergenic region via quantitative PCR (qPCR).
- Quantitative RT-PCR (qRT-PCR): Extract total RNA from control and NDR1-deficient cells. Measure the relative mRNA expression levels of miR146a primary transcript and mature form, as well as ISGs.
- Western Blotting: Analyze protein lysates from control and NDR1-manipulated cells with or without viral infection (e.g., using poly(I:C)) or IFN stimulation. Probe for STAT1, phospho-STAT1, and other ISG proteins to assess translation and pathway activation.
- Luciferase Reporter Assay: Co-transfect cells with an IFN-stimulated response element (ISRE) luciferase reporter plasmid and NDR1 expression vectors (wild-type and kinase-dead). Measure luciferase activity after IFN stimulation to test kinase activity dependence.
Protocol: Investigating NDR2's Role in RIG-I Activation
- Key Study: NDR2 promotes TRIM25-mediated K63-linked ubiquitination of RIG-I [56].
- Objective: To elucidate the mechanism by which NDR2 facilitates RIG-I activation.
- Methodology:
- Co-Immunoprecipitation (Co-IP) and Western Blotting:
- Validation of Interactions: Transfect cells with plasmids for NDR2, RIG-I, and TRIM25. Immunoprecipitate one protein (e.g., NDR2) and probe the blot for the others (e.g., RIG-I and TRIM25) to confirm direct association.
- Ubiquitination Assay: Co-transfect cells with plasmids for RIG-I, TRIM25, ubiquitin, and NDR2 (or control). Immunoprecipitate RIG-I and probe the membrane with an antibody specific for K63-linked ubiquitin chains to assess the level of RIG-I ubiquitination.
- Viral Infection Models: Infect control and Ndr2 knockout macrophages with RNA viruses (e.g., Respiratory Syncytial Virus). Measure the production of IFN-β and pro-inflammatory cytokines using ELISA, and viral replication via plaque assay.
- In Vivo Validation: Use conditional knockout mice (e.g., LysmâºNDR2f/f) to study the cell-type-specific role of NDR2 in antiviral defense against viral challenge.
- Co-Immunoprecipitation (Co-IP) and Western Blotting:
Signaling Pathways Visualization
The Scientist's Toolkit: Key Research Reagents
Table 2: Essential Reagents for Studying NDR1 and NDR2 in Innate Immunity
| Reagent / Tool | Function / Application | Key Example in Context |
|---|---|---|
| siRNA/shRNA | Gene knockdown to study loss-of-function phenotypes. | Knockdown of NDR2 with siRNA increased CpG-induced IL-6 secretion, suggesting functional similarity to NDR1 in TLR9 signaling [14]. |
| Conditional Knockout Mice | In vivo validation of cell-type-specific functions. | LysmâºNDR2f/f mice showed an impaired antiviral immune response [56]. Stk38-deficient mice were more susceptible to bacterial sepsis [14]. |
| Kinase-Inactive Mutants | Differentiating between kinase-dependent and scaffolding functions. | Overexpression of NDR2 kinase-inactive mutants still potentiated the antiviral response, indicating a scaffolding role [56]. |
| Co-IP & Western Blotting | Protein-protein interaction and post-translational modification analysis. | Used to demonstrate the association between NDR2, RIG-I, and TRIM25, and to measure K63-linked ubiquitination of RIG-I [56]. |
| Luciferase Reporter Assays | Measuring transcriptional activity of signaling pathways. | An IFN-β promoter luciferase reporter can be used to assess the impact of NDR1/2 on RIG-I signaling output [58]. |
| Viral Pathogen Assays | Functional validation of antiviral activity. | Respiratory Syncytial Virus (RSV) and other RNA viruses used to infect NDR2-manipulated macrophages, followed by viral titer measurement (plaque assay) [56]. |
NDR1 and NDR2, despite their structural similarity, have evolved non-redundant, specialized roles in innate immunity dictated by their distinct subcellular localizations. NDR1 operates as a nuclear transcriptional regulator, fine-tuning the immune response by controlling the expression of key immune modulators like miR146a and STAT1. In contrast, NDR2 functions in the cytoplasm as a direct facilitator of the RIG-I/TRIM25 complex, acting as a critical scaffold to promote the ubiquitination and activation of RIG-I. This clear functional divergence underscores the complexity of immune regulation and highlights the potential of targeting these kinases in a context-specific manner for therapeutic intervention in infectious and inflammatory diseases.
NDR2's Regulation of Integrin-Dependent Synaptic Plasticity vs. NDR1's Emerging Nuclear Roles
The Nuclear Dbf2-related (NDR) serine/threonine kinases, NDR1 (STK38) and NDR2 (STK38L), represent two highly homologous enzymes with 87% amino acid sequence identity that have evolved distinct functional specializations within the mammalian nervous system [6] [4]. Despite their structural similarity, these kinases exhibit fundamentally different subcellular localizations: NDR1 is predominantly nuclear, while NDR2 is excluded from the nucleus and displays a punctate cytoplasmic distribution [6] [4]. This fundamental difference in localization underpins their specialized roles in neuronal function. NDR2 has emerged as a critical regulator of integrin-dependent synaptic plasticity, dendritic branching, and neurite outgrowth through cytoplasmic signaling mechanisms [60] [61]. Meanwhile, NDR1's nuclear localization enables distinct functions in gene regulation and cellular homeostasis [3] [8]. This comparison guide examines the experimental evidence defining their specialized neuronal functions, providing researchers with methodological insights and resource information to advance investigations into these important signaling kinases.
Table 1: Fundamental Characteristics of NDR1 and NDR2 Kinases
| Feature | NDR1 (STK38) | NDR2 (STK38L) |
|---|---|---|
| Primary Localization | Nuclear [3] [8] | Cytoplasmic/Punctate [6] [4] |
| Expression in Adult Mouse CNS | Absent [60] | Principal NDR kinase [60] |
| C-terminal Targeting Signal | Ala-Lys [4] | Gly-Lys-Leu (Peroxisomal targeting) [4] |
| Key Neuronal Functions | Cell cycle regulation, apoptosis [3] | Synaptic plasticity, dendritic branching, neurite outgrowth [60] [61] |
| Hippo Pathway Involvement | Core component [3] [62] | Core component [60] [62] |
Subcellular Localization Mechanisms: Directing Functional Specialization
The divergent subcellular localization of NDR1 and NDR2 represents a critical determinant of their functional specialization. NDR1 contains a nuclear localization signal (residues 265-276) that facilitates its predominant nuclear distribution [8]. In contrast, NDR2 exhibits a punctate cytoplasmic localization despite possessing a nearly identical NLS sequence with only a single conservative amino acid change [8] [6]. Recent research has revealed that NDR2's distinctive distribution is mediated by a C-terminal peroxisomal targeting signal type 1 (PTS1)-like sequence (Gly-Lys-Leu) that is absent in NDR1 (which terminates in Ala-Lys) [4]. This GKL motif enables NDR2 to bind the PTS1 receptor Pex5p, directing its localization to peroxisomes—single-membrane organelles crucial for metabolic functions and signaling [4]. Mutation of the C-terminal leucine in NDR2 (NDR2(ΔL)) results in diffuse cytoplasmic distribution similar to NDR1, confirming the critical nature of this targeting motif [4].
Diagram 1: Differential localization mechanisms of NDR1 and NDR2. NDR1 enters the nucleus via its NLS, while NDR2 localizes to peroxisomes through Pex5p recognition of its C-terminal GKL motif.
This fundamental difference in subcellular targeting directs each kinase to distinct microenvironments with specialized signaling components, ultimately determining their neuronal functions. The peroxisomal localization of NDR2 appears crucial for its role in primary cilium formation, as wild-type NDR2—but not the peroxisome-non-targeting NDR2(ΔL) mutant—can rescue ciliogenesis defects in NDR2-deficient cells [4].
NDR2 in Integrin-Dependent Synaptic Plasticity: Mechanisms and Experimental Evidence
Molecular Mechanisms of NDR2 in Synaptic Regulation
NDR2 serves as a critical regulator of synaptic formation and plasticity through integrin-mediated signaling pathways in the hippocampal CA1 region [60]. The molecular mechanism involves NDR2 phosphorylation of Filamin A at serine 2152, which promotes Filamin A dissociation from the β1 integrin tail domain [60]. This dissociation enables activators Talin and Kindlin to associate with β1 integrins, facilitating full integrin activation through phosphorylation of the T788/789 motif—a process essential for integrin signaling [60]. In NDR2 null mutant mice, this pathway is disrupted, leading to reduced T788/789 phosphorylated β1 integrin expression at synaptic sites, decreased synaptic density, and impaired long-term potentiation (LTP) at Schaffer collateral/commissural fiber-CA1 pyramidal cell synapses [60]. Significantly, this LTP impairment can be rescued by integrin activation with arginine-glycine-aspartate (RGD)-containing peptides, demonstrating the crucial position of NDR2 upstream of integrin activation in the synaptic plasticity pathway [60].
Diagram 2: NDR2 regulates synaptic plasticity through integrin signaling. In wild-type neurons (colored), NDR2 phosphorylates Filamin A, leading to integrin activation and normal LTP. Under NDR2 deficiency (gray), this pathway is disrupted, impairing synaptic plasticity.
Beyond synaptic plasticity, NDR2 determines substrate specificity for neurite extension through regulation of α1 integrin expression and trafficking [61]. PC12 cells overexpressing NDR2 show enhanced neurite growth on poly-D-lysine substrates but fail to exhibit the α1β1 integrin-mediated enhancement of neurite growth on collagen IV substrate seen in control cells [61]. This suggests NDR2 regulates both the activation state and subtype-specific trafficking of integrins to control neurite outgrowth in an extracellular matrix-dependent manner.
Experimental Models and Methodologies for Studying NDR2
Research into NDR2 function has employed sophisticated genetic and physiological approaches. Key studies utilize constitutive NDR2 null mutant mice (Stk38lGt(RRT116)byg) generated through gene-trap technology in ES cell line E14TG2a, back-crossed for more than 12 generations to C57Bl/6 backgrounds [60]. Electrophysiological assessment of synaptic plasticity involves measuring field excitatory postsynaptic potentials (fEPSPs) in hippocampal slices following high-frequency stimulation (HFS) to induce LTP [60]. Integin rescue experiments employ RGD-containing peptides (e.g., Gly-Arg-Gly-Asp-Ser-Pro) applied to hippocampal slices to bypass NDR2 deficiency and directly activate integrins [60].
Behavioral analysis includes Morris water maze and water cross maze tests to assess spatial memory deficits in NDR2-deficient mice [60]. In vitro studies utilize primary hippocampal neurons and PC12 cell lines stably expressing EGFP-tagged NDR2 to examine neurite outgrowth and dendritic branching on various ECM substrates including poly-D-lysine, collagen IV, fibronectin, and laminin [61]. Quantitative analysis of neurite growth involves counting cells with neurites longer than 100μm following NGF-induced differentiation [61].
Table 2: Key Experimental Findings on NDR2 in Synaptic Plasticity
| Experimental Approach | Key Finding | Quantitative Data | Citation |
|---|---|---|---|
| NDR2 KO mice hippocampal LTP | Reduced LTP in CA1 synapses | Restored to normal levels with RGD peptide | [60] |
| Phosphorylated β1 integrin staining | Reduced pT788/789 β1 integrin in KO | Significant decrease at synaptic sites | [60] [61] |
| Synaptic density measurement | Decreased synaptic density in KO | Significant reduction in hippocampal CA1 | [60] |
| Spatial memory testing (MWM) | Mild spatial memory deficits | Impaired performance in KO mice | [60] |
| Neurite outgrowth assays | Altered growth on collagen IV | NDR2 OE cells lost α1β1-mediated enhancement | [61] |
NDR1's Nuclear Functions: Emerging Roles in Neuronal Regulation
While NDR2 functions predominantly in cytoplasmic signaling, NDR1's nuclear localization enables distinct regulatory roles. In innate immunity and inflammation—processes increasingly recognized as important in neuronal function—NDR1 serves as a negative regulator of TLR9-mediated immune response in macrophages [3]. NDR1 achieves this by binding the ubiquitin E3 ligase Smurf1, promoting Smurf1-mediated ubiquitination and degradation of MEKK2, which is essential for CpG-induced ERK1/2 activation and subsequent production of TNF-α and IL-6 [3]. This regulatory function has implications for neuroinflammatory processes, with Stk38-deficient mice showing higher levels of pro-inflammatory cytokines and increased mortality following E. coli infection or cecal ligation and puncture-induced polymicrobial sepsis [3].
NDR1 also functions as a transcriptional regulator by binding to the intergenic region of miR146a, dampening its transcription to promote STAT1 translation and enhance antiviral immune response independently of its kinase activity [3]. This nuclear function in gene regulation represents a distinct mechanism from NDR2's cytoplasmic signaling and illustrates how subcellular localization dictates functional specialization.
Additionally, NDR1 and NDR2 play coordinated roles in neuronal membrane trafficking and protein homeostasis, with double knockout of both Ndr1 and neuron-specific Ndr2 causing neurodegeneration with progressive accumulation of p62 and ubiquitinated proteins [63]. Neurons from double knockout mice show reductions in autophagosome numbers and altered endosomal compartments, indicating that both kinases contribute to maintaining neuronal protein homeostasis, albeit through potentially different mechanisms [63].
Comparative Experimental Approaches: Methodological Considerations
Key Methodologies for Investigating NDR1 vs. NDR2 Functions
The distinct functions of NDR1 and NDR2 necessitate specialized experimental approaches. For localization studies, researchers employ N-terminally tagged YFP-NDR1 and YFP-NDR2 constructs transfected into cell lines (e.g., RPE1, HeLa, COS-7) followed by immunostaining with organelle-specific markers (catalase for peroxisomes, EEA1 for early endosomes, GM130 for Golgi) [4]. Subcellular fractionation and iodixanol density gradient ultracentrifugation provide biochemical confirmation of localization, with NDR2 co-sedimenting with peroxisomal protein Pex14p [4].
Functional studies of NDR2 in neurite outgrowth utilize PC12 cell differentiation assays on various ECM substrates (poly-D-lysine, collagen I/IV, fibronectin, laminin) with NGF stimulation [61]. Quantitative analysis measures percentage of cells with neurites >100μm after 4 days of differentiation [61]. Integrin activation states are assessed using phosphorylation-specific antibodies against β1 integrin pT788/789 [61]. For dendritic branching studies, primary hippocampal neurons are cultured from C57BL/6 mice on different substrates with analysis of dendritic complexity [61].
Electrophysiological assessment of NDR2 function involves hippocampal slice preparations from NDR2 KO mice with field potential recordings in CA1 region following Schaffer collateral stimulation [60]. LTP is induced using high-frequency stimulation (e.g., 100Hz tetanus), with integrin rescue experiments employing RGD-containing peptides in the perfusion medium [60].
Research Reagent Solutions
Table 3: Essential Research Reagents for NDR Kinase Investigation
| Reagent/Cell Line | Specific Example | Research Application | Function |
|---|---|---|---|
| Animal Models | Stk38lGt(RRT116)byg mice (NDR2 KO) | Synaptic plasticity, behavior | Constitutive NDR2 knockout [60] |
| Cell Lines | PC12 cells stably expressing EGFP-NDR2 | Neurite outgrowth, integrin trafficking | Neuronal differentiation model [61] |
| Antibodies | pT788/789 β1 integrin (Abcam ab5189) | Integrin activation assays | Detects activated β1 integrin [61] |
| Peptide Reagents | RGD-containing (GRGDSP) | Integrin rescue experiments | Activates integrins independently of NDR2 [60] |
| Expression Vectors | YFP-NDR1/NDR2, CFP-SKL | Subcellular localization studies | Visualize kinase localization and peroxisomal targeting [4] |
| ECM Substrates | Collagen IV, laminin, fibronectin | Neurite growth specificity assays | Test integrin-substrate interactions [61] |
NDR1 and NDR2 kinase represent a compelling example of functional specialization through subcellular localization. While sharing significant structural homology, their distinct targeting—nuclear for NDR1 versus cytoplasmic/peroxisomal for NDR2—directs them to fundamentally different cellular processes. NDR2 has emerged as a master regulator of integrin-dependent synaptic plasticity, controlling dendritic branching, neurite outgrowth, and hippocampal LTP through phosphorylation of Filamin A and subsequent activation of β1 integrin signaling [60] [61]. Meanwhile, NDR1's nuclear localization enables gene regulatory functions in inflammatory response and potentially other nuclear processes [3]. Both kinases play coordinated roles in maintaining neuronal health, with combined deficiency leading to neurodegeneration through disrupted autophagy and protein homeostasis [63].
For researchers investigating these kinases, the experimental approaches and reagents outlined here provide essential methodological guidance. The continuing elucidation of NDR1 and NDR2 functions offers promising insights into neuronal development, plasticity, and disease mechanisms, with potential applications in therapeutic development for neurological disorders.
The Hippo signaling pathway is an evolutionarily conserved system that functions as a critical regulator of organ size, tissue homeostasis, and tumor suppression by precisely controlling cell proliferation, apoptosis, and stem cell self-renewal [64] [65]. This pathway has garnered significant scientific interest due to its dual role in physiological regulation and pathological processes, particularly in cancer development and progression. At the heart of the Hippo pathway lies a sophisticated regulatory mechanism that governs the subcellular localization and transcriptional activity of its downstream effectors, Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ). The precise control of YAP/TAZ partitioning between cytoplasmic sequestration and nuclear translocation represents a fundamental biological process that determines transcriptional output and ultimately directs cellular fate decisions [66] [67].
The clinical relevance of this pathway is substantial, as dysregulation of Hippo signaling and consequent aberrant YAP/TAZ activation are frequently observed in diverse human cancers, including non-small cell lung cancer (NSCLC), colorectal cancer, breast cancer, and hepatocellular carcinoma [68] [69] [65]. Understanding the molecular mechanisms that dictate the differential regulation of YAP/TAZ localization and activity provides critical insights for developing novel therapeutic strategies targeting this pathway in cancer and other diseases. This review systematically examines the complex regulatory networks controlling cytoplasmic YAP/TAZ sequestration and nuclear transcriptional output, with particular emphasis on emerging concepts in the field, including biomolecular condensates and their implications for Hippo pathway-directed therapeutics.
Core Hippo Pathway Components and Regulatory Mechanisms
The Canonical Kinase Cascade: From Membrane to Nucleus
The core Hippo pathway consists of a highly conserved kinase cascade that transmits inhibitory signals from the cell membrane to the nucleus. Key upstream kinases include Mammalian STE20-like protein kinases 1/2 (MST1/2), which complex with their adaptor protein Salvador homolog 1 (SAV1). This kinase complex then phosphorylates and activates the downstream kinases Large Tumor Suppressor Kinases 1/2 (LATS1/2) in conjunction with their scaffold proteins MOB kinase activator 1A/B (MOB1A/B) [64] [65]. The fully activated LATS1/2 directly phosphorylates the transcriptional coactivators YAP and TAZ, marking them for either cytoplasmic retention or proteasomal degradation [69].
The regulation of this kinase cascade involves multiple upstream inputs, including cell polarity proteins, mechanical cues from the extracellular matrix (ECM), cell density signals, soluble factors, and various cellular stress signals [64]. These diverse inputs are integrated through the core kinase cascade to precisely modulate YAP/TAZ activity according to cellular context and environmental conditions, enabling appropriate physiological responses during development, tissue repair, and homeostatic maintenance.
YAP/TAZ: Central Effectors of Hippo Signaling
YAP and TAZ serve as the primary effectors of the Hippo pathway, shuttling between cytoplasmic and nuclear compartments to regulate gene expression programs. When the Hippo pathway is activated, LATS1/2 phosphorylates YAP at multiple serine residues (primarily Ser127 in YAP), creating binding sites for 14-3-3 proteins that sequester YAP/TAZ in the cytoplasm [69] [67]. Alternatively, phosphorylated YAP/TAZ can undergo ubiquitination mediated by casein kinase 1 (CK1) and β-transducin repeat-containing protein (β-TrCP), leading to proteasomal degradation [69].
When Hippo signaling is inactive, dephosphorylated YAP/TAZ translocates to the nucleus where it interacts primarily with Transcriptional Enhanced Associate Domain (TEAD) family transcription factors (TEAD1-4) to activate target gene expression [66] [70]. YAP/TAZ lack intrinsic DNA-binding domains and therefore depend entirely on transcription factors like TEADs to target specific genomic loci [67]. The YAP/TAZ-TEAD complex drives the expression of genes involved in cell proliferation (e.g., CTGF, CYR61), survival, migration, and stemness maintenance [66] [65].
Table 1: Core Components of the Hippo Signaling Pathway
| Component Category | Key Molecules | Primary Functions |
|---|---|---|
| Upstream Kinases | MST1/2, MAP4Ks | Initiate kinase cascade; phosphorylate LATS1/2 |
| Adaptor Proteins | SAV1, MOB1A/B | Scaffold functions; facilitate kinase interactions |
| Core Kinases | LATS1/2 | Directly phosphorylate YAP/TAZ |
| Transcriptional Effectors | YAP, TAZ | Transcriptional co-activators; shuttle between cytoplasm and nucleus |
| DNA-Binding Partners | TEAD1-4 | Primary transcription factors partnering with YAP/TAZ |
| Negative Regulators | VGLL4 | Competes with YAP for TEAD binding; represses transcription |
Cytoplasmic Sequestration Mechanisms of YAP/TAZ
Phosphorylation-Dependent Sequestration
The primary mechanism for cytoplasmic retention of YAP/TAZ involves LATS1/2-mediated phosphorylation, which occurs when the Hippo pathway is activated in response to various upstream signals. Phosphorylated YAP/TAZ binds to 14-3-3 chaperone proteins through specific phospho-serine motifs, effectively trapping them in the cytoplasmic compartment and preventing nuclear translocation [69] [67]. This molecular interaction represents a critical control point in Hippo pathway regulation, as it directly determines the proportion of YAP/TAZ molecules capable of accessing the nuclear compartment and activating transcription.
Beyond 14-3-3 binding, phosphorylation also targets YAP/TAZ for proteasomal degradation through a well-defined ubiquitination pathway. Phosphorylated YAP/TAZ is recognized by the E3 ubiquitin ligase complex containing β-TrCP, leading to polyubiquitination and subsequent degradation by the proteasome [69]. This dual mechanism of cytoplasmic sequestration—through both physical retention and targeted degradation—ensures tight control over YAP/TAZ activity and provides a robust system for preventing aberrant transcriptional activation under normal physiological conditions.
Regulation by Cell Polarity and Adhesion Complexes
Cell polarity proteins and adhesion complexes play crucial roles in regulating YAP/TAZ localization and activity. Proteins located at apical domains, such as Merlin/NF2, function as upstream activators of the Hippo pathway by facilitating the recruitment and activation of core kinase components [64]. Similarly, the Angiomotin (AMOT) family proteins, essential components of tight junctions, act as scaffolds that promote LATS1/2-mediated phosphorylation of YAP/TAZ [64] [65]. These spatial constraints ensure that YAP/TAZ activity is appropriately suppressed in polarized epithelial tissues.
At adherens junctions, α-Catenin physically interacts with YAP and contributes to its cytoplasmic retention independently of phosphorylation, providing an additional layer of regulation [64]. This mechanism links cell-cell contact to YAP/TAZ inhibition, partly explaining the phenomenon of contact inhibition observed in densely cultured cells. The integration of structural information from cell junctions with Hippo signaling creates a sophisticated system for coordinating tissue architecture with proliferative signals, ensuring proper tissue organization and preventing overgrowth.
Mechanical Regulation and Extracellular Matrix Influences
The extracellular matrix (ECM) composition and stiffness significantly influence YAP/TAZ localization through mechanotransduction pathways. On stiff substrates that promote cell spreading, integrin-mediated signaling leads to activation of Src family kinases and Focal Adhesion Kinase (FAK), which inactivate the Hippo pathway and promote YAP/TAZ nuclear localization [71] [66]. Conversely, soft substrates or reduced cell spreading promote Hippo pathway activation and cytoplasmic YAP/TAZ retention [66].
The actin cytoskeleton serves as a central mediator of this mechanical regulation. Increased actin polymerization and contractility, often driven by Rho GTPases and ROCK signaling, correlate with YAP/TAZ activation and nuclear translocation [66]. This mechanosensitive regulation allows cells to integrate physical cues from their microenvironment with transcriptional programs, enabling appropriate responses to changes in tissue stiffness, a particularly relevant mechanism in fibrotic diseases and cancer progression where ECM remodeling frequently occurs.
Nuclear Translocation and Transcriptional Activation
TEAD-Dependent Transcriptional Mechanisms
Upon nuclear translocation, YAP/TAZ primarily form complexes with TEAD transcription factors (TEAD1-4) to regulate gene expression. The interaction between YAP/TAZ and TEADs displaces transcriptional repressors such as VGLL4, converting TEADs from transcriptional repressors to activators [66] [70]. This molecular switch activates the expression of a broad array of target genes involved in cell proliferation (e.g., AXL, BIRC5), survival, migration, and epithelial-mesenchymal transition [66] [69].
Recent research has revealed that the majority (approximately 91%) of YAP/TAZ/TEAD binding occurs at distal enhancer regions rather than promoter regions, highlighting the importance of enhancer regulation in Hippo pathway-mediated transcription [67]. These enhancer elements often display characteristics of "super-enhancers"—large regulatory regions heavily loaded with coactivators, mediator complexes, and activating histone marks such as H3K27ac [67]. YAP recruits the CDK9 kinase to these enhancer regions, which promotes the release of paused RNA Polymerase II and enables productive transcriptional elongation of target genes [67].
Biomolecular Condensates and Phase Separation
Emerging evidence indicates that liquid-liquid phase separation (LLPS) plays a crucial role in organizing Hippo pathway components within the nucleus. Recent studies demonstrate that TEAD transcription factors possess an intrinsic capacity to form biomolecular condensates that function as nuclear signaling hubs [70]. Rather than driving phase separation themselves, endogenous YAP/TAZ proteins act as "client" proteins that partition into these pre-formed TEAD condensates [70].
These TEAD condensates serve as organizational centers that dynamically concentrate transcriptionally active YAP along with other regulators of transcriptional activation, forming "transcription factories" within the nucleus [70]. This compartmentalization through phase separation potentially explains how the Hippo pathway can achieve specific transcriptional responses despite the relatively ubiquitous expression of its components. The discovery of this mechanism opens new avenues for therapeutic intervention, as disrupting these condensates could offer a novel approach to modulating Hippo pathway output in disease states.
Context-Specific Transcriptional Partnerships
While TEADs represent the primary partners for YAP/TAZ, these transcriptional coactivators can also interact with additional transcription factors to achieve context-specific gene regulation. YAP/TAZ have been reported to form complexes with AP-1 factors, SMADs (in TGF-β signaling), p73, RUNX1/2, ERBB4, and TBX5 [66] [67]. These alternative partnerships allow the Hippo pathway to integrate with other signaling networks and tailor transcriptional outputs according to cellular context and lineage-specific requirements.
The specific transcriptional programs activated by YAP/TAZ vary significantly across different tissue types and cellular states. In progenitor cells, YAP/TAZ promote stemness and self-renewal, while in differentiated tissues, they can drive regenerative responses following injury [67]. This contextual plasticity enables the Hippo pathway to fulfill diverse functions in development, homeostasis, and repair, but also contributes to its pathogenic roles in cancer when dysregulated.
Experimental Approaches for Studying Hippo Regulation
Methodologies for Assessing YAP/TAZ Localization and Activity
Investigating the subcellular localization and functional status of YAP/TAZ requires specialized experimental approaches that can capture their dynamic regulation. Immunofluorescence microscopy serves as the primary method for visualizing YAP/TAZ localization, with nuclear accumulation indicating activation and cytoplasmic localization indicating inhibition [71] [66]. This technique can be combined with pharmacological inhibitors or genetic manipulations to dissect regulatory mechanisms.
Fractionation studies followed by Western blotting provide a quantitative complement to microscopic localization assays, allowing precise measurement of YAP/TAZ distribution between nuclear and cytoplasmic compartments [71]. To assess functional activity, researchers employ reporter gene assays using TEAD-responsive promoters (e.g., 8xGTIIC-luciferase) to measure transcriptional output directly [70]. Additionally, co-immunoprecipitation experiments can validate physical interactions between YAP/TAZ and their binding partners such as TEADs or LATS1/2 [71] [70].
Table 2: Key Experimental Methods for Hippo Pathway Analysis
| Method Category | Specific Techniques | Primary Applications | Key Insights Generated |
|---|---|---|---|
| Localization Analysis | Immunofluorescence, Cellular fractionation + Western blot | Determine YAP/TAZ subcellular distribution | Correlation of nuclear localization with pathway activation; response to mechanical cues |
| Functional Assays | TEAD-luciferase reporter, RNA-seq, ChIP-seq | Measure transcriptional activity; identify target genes | Assessment of pathway output; identification of direct vs. indirect targets |
| Interaction Studies | Co-immunoprecipitation, Proximity ligation | Protein-protein interactions; complex formation | Validation of YAP-TEAD, YAP-LATS interactions; drug disruption studies |
| Genetic Manipulation | CRISPR-Cas9, siRNA/shRNA | Loss-of-function studies; pathway dissection | Identification of essential components; functional hierarchies |
| Metabolic Studies | Seahorse Analyzer, Metabolomics | Bioenergetic profiling; metabolic dependencies | Link between Hippo signaling and metabolic reprogramming in cancer |
Investigating Pathway Regulation Using Genetic and Pharmacological Tools
Genetic approaches have been instrumental in delineating Hippo pathway components and their functional relationships. RNA interference (siRNA/shRNA) and CRISPR-Cas9 technologies enable targeted depletion of specific pathway components, allowing researchers to establish functional hierarchies and identify essential regulators [12]. For example, CRISPR-Cas9-mediated knockout of Ndr2/Stk38l in microglial cells revealed its specific role in metabolic adaptation under high-glucose conditions, distinct from its paralog Ndr1 [12].
Pharmacological inhibitors provide complementary tools for acute pathway manipulation. Several small molecule inhibitors targeting different nodes of the Hippo pathway have been developed, including verteporfin (disrupts YAP-TEAD interaction) and central pocket inhibitors that target the palmitoylation pocket of TEADs (e.g., VT3989, IK-930, IAG933) [68] [70]. These compounds have shown promise in preclinical cancer models and some have advanced to clinical trials, highlighting their utility as both research tools and therapeutic agents.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Hippo Pathway Investigation
| Reagent Category | Specific Examples | Research Applications | Experimental Notes |
|---|---|---|---|
| Cell Line Models | Caco-2 (colorectal), SF268 (glioblastoma), MDA-MB-231 (breast) | Context-specific pathway analysis; drug screening | Different lines show varying baseline Hippo activity; select based on research focus |
| Antibodies | YAP/TAZ (Cell Signaling #8418), Pan-TEAD, p-YAP (Ser127) | Immunofluorescence, Western blot, Co-IP | Phospho-specific antibodies crucial for assessing pathway activity status |
| Inhibitors | Verteporfin (YAP-TEAD), VT3989 (TEAD palmitoylation) | Pathway inhibition studies; therapeutic testing | Different mechanisms of action; varying efficacy across cell types |
| Expression Plasmids | YAP(5SA) (constitutively active), TEAD mutants | Gain-of-function studies; mechanism dissection | Constitutive active YAP lacks LATS phosphorylation sites |
| CRISPR Tools | sgRNAs targeting LATS1/2, NDR1/2, TEAD1-4 | Genetic knockout; functional validation | Consider functional redundancy between paralogs (e.g., LATS1/2) |
| Reporters | 8xGTIIC-luciferase, CTGF-luciferase | Transcriptional activity measurement | High-throughput screening for modulators |
Visualization of Hippo Pathway Regulation
The core regulatory mechanism of the Hippo pathway, encompassing both cytoplasmic sequestration and nuclear activation of YAP/TAZ, can be visualized through the following integrated signaling network:
Visualization 1: Integrated Hippo Signaling Pathway Regulation. This diagram illustrates the core Hippo kinase cascade leading to cytoplasmic YAP/TAZ sequestration (red) and the nuclear transcriptional activation mechanism (blue), highlighting key regulatory nodes and the newly discovered role of TEAD biomolecular condensates.
The process of TEAD biomolecular condensate formation and YAP/TAZ recruitment can be specifically visualized as follows:
Visualization 2: TEAD Biomolecular Condensate Formation and Function. This diagram details the process of TEAD phase separation, subsequent YAP/TAZ recruitment, and formation of transcriptional "factories," including potential therapeutic intervention points.
Concluding Perspectives and Future Directions
The differential regulation of Hippo pathway signaling through cytoplasmic sequestration and nuclear translocation of YAP/TAZ represents a sophisticated biological control mechanism with profound implications for both normal physiology and disease pathogenesis. The emerging understanding of biomolecular condensates and their role in organizing Hippo pathway components within the nucleus adds a new layer of complexity to our conceptual framework [70]. This discovery not only advances fundamental knowledge of cellular organization but also presents novel opportunities for therapeutic intervention in cancers characterized by Hippo pathway dysregulation.
Future research directions should focus on elucidating the context-specific determinants of YAP/TAZ transcriptional outputs, particularly how different tissue environments and cellular states influence pathway activity. The development of more selective TEAD inhibitors that can discriminate between different TEAD paralogs or disrupt specific protein-protein interactions holds promise for improved therapeutic efficacy with reduced off-target effects [70]. Additionally, exploring the crosstalk between Hippo signaling and other oncogenic pathways may reveal combination therapies that can overcome resistance mechanisms and improve patient outcomes across multiple cancer types.
As single-cell technologies and spatial transcriptomics continue to advance, researchers will gain unprecedented resolution into the heterogeneity of Hippo pathway activity within complex tissues and tumor microenvironments [67]. These insights will undoubtedly refine our understanding of how cytoplasmic-nuclear partitioning of YAP/TAZ is regulated across different cellular contexts and how this precise regulation can be therapeutically manipulated for cancer treatment and regenerative medicine applications.
The NDR (Nuclear Dbf2-Related) kinase family, comprising NDR1 and NDR2, represents a crucial subclass of serine/threonine kinases within the Hippo signaling pathway. Despite their significant structural similarity (87% amino acid sequence identity), these kinases exhibit fundamentally distinct subcellular localizations that dictate their specialized biological functions [4] [6] [10]. NDR1 primarily localizes to the nucleus, while NDR2 demonstrates a punctate cytoplasmic distribution, specifically targeting peroxisomes through a C-terminal peroxisome-targeting signal (PTS1) type sequence [4] [6]. This localization dichotomy establishes a functional specialization where NDR2 plays unique roles in cellular adaptation to metabolic stress, particularly in immune cells such as microglia exposed to high-glucose environments [72] [12].
In diabetic retinopathy, a major complication of diabetes characterized by chronic retinal inflammation, microglial cells undergo metabolic reprogramming that drives disease progression [72] [12]. Understanding how NDR2 regulates microglial adaptation to high glucose conditions provides critical insights into the neuroinflammatory processes underlying this vision-threatening condition and reveals potential therapeutic targets for mitigating retinal inflammation [72].
Comparative Analysis: NDR1 versus NDR2 Structural and Functional Differences
Table 1: Fundamental Differences Between NDR1 and NDR2 Kinases
| Characteristic | NDR1 (STK38) | NDR2 (STK38L) |
|---|---|---|
| Subcellular Localization | Diffuse nuclear and cytoplasmic distribution [4] [6] | Punctate cytoplasmic distribution; peroxisomal localization [4] |
| C-terminal Targeting Signal | Ala-Lys [4] | Gly-Lys-Leu (GKL) - functions as PTS1 [4] |
| Pex5p Binding | No binding [4] | Direct binding to PTS1 receptor [4] |
| Primary Cilium Formation | No significant role [4] | Critical regulator of ciliogenesis [4] |
| Microglial Metabolic Regulation | Not implicated [12] | Key regulator of glucose-dependent adaptation [72] [12] |
The functional specialization between NDR1 and NDR2 stems primarily from their distinct C-terminal sequences, which dictate differential subcellular targeting [4] [10]. NDR2 contains a C-terminal Gly-Lys-Leu (GKL) sequence that functions as a peroxisome-targeting signal (PTS1), enabling its specific localization to peroxisomes through interaction with the PTS1 receptor Pex5p [4]. In contrast, NDR1 terminates in Ala-Lys and exhibits diffuse cellular distribution without specific organelle association [4] [6]. This fundamental difference in localization underlies their specialized functions, with NDR2 playing a unique role in regulating metabolic adaptation and inflammatory responses in microglial cells under high-glucose conditions [72] [12].
NDR2 in Microglial Adaptation to High Glucose: Experimental Findings
NDR2 Expression Dynamics Under Metabolic Stress
Table 2: NDR2 Protein and mRNA Expression in BV-2 Microglial Cells Under High-Glucose Conditions
| Experimental Condition | NDR2 Protein Level | NDR2 mRNA Level | Experimental Details |
|---|---|---|---|
| 7-hour HG exposure | Significant increase (CT: 24.0 ± 4.4 a.u.; HG: 83.0 ± 19.1 a.u.) [12] | No significant change (HG: 80.0 ± 0.1% of CT) [12] | 30.5 mM glucose vs. 5.5 mM normal glucose [12] |
| 12-hour HG exposure | Significant increase (CT: 26.1 ± 6.9 a.u.; HG: 64.2 ± 10.1 a.u.) [12] | Trend toward increase (HG: 160.3 ± 34.0% of CT, p=0.097) [12] | Two 4-hour HG periods separated by 4-hour normal glucose [12] |
Exposure to high-glucose conditions induces significant upregulation of NDR2 protein expression in microglial cells without corresponding increases in mRNA levels, suggesting post-translational regulation mechanisms [12]. This response indicates that NDR2 functions as a stress-adaptive kinase that helps microglia cope with metabolic challenges associated with diabetic conditions [72] [12].
Functional Consequences of NDR2 Downregulation
Table 3: Functional Deficits in NDR2-Downregulated Microglial Cells
| Functional Parameter | Effect of NDR2 Downregulation | Significance |
|---|---|---|
| Mitochondrial Respiration | Impaired [72] [12] | Reduced metabolic flexibility and defective stress adaptation [72] |
| Phagocytic Capacity | Reduced [72] [12] | Impaired cytoskeletal dynamics and debris clearance [72] |
| Migratory Capacity | Reduced [72] [12] | Disrupted surveillance and response capabilities [72] |
| Pro-inflammatory Cytokines | Elevated (IL-6, TNF, IL-17, IL-12p70) [72] [12] | Enhanced inflammatory response even under normal glucose [72] |
CRISPR-Cas9-mediated partial knockout of Ndr2/Stk38l in BV-2 mouse microglial cells reveals its critical role in maintaining metabolic flexibility and regulating inflammatory responses [72] [12]. Microglia with impaired NDR2 function exhibit widespread functional deficits despite normal glucose conditions, indicating NDR2's fundamental role in microglial homeostasis independent of acute metabolic stress [72].
Experimental Methodologies for Investigating NDR2 Function
CRISPR-Cas9-Mediated NDR2 Downregulation
Protocol: Early passage BV-2 cells (passage 7) were transfected via lipofectamine with an all-in-one plasmid containing sgRNA targeting exon 7 of the Ndr2 gene to disrupt NDR2 expression [12]. The partial knockout efficiency was validated through Western blot analysis and functional assays [12].
Application: This approach enables specific investigation of NDR2 function without complete kinase ablation, modeling the partial dysfunction that may occur in pathological conditions [12].
Metabolic and Functional Assays
Mitochondrial Respiration Assessment: Using Seahorse Analyzer technology, researchers measured oxygen consumption rates (OCR) in control and NDR2-downregulated microglial cells under both normal and high-glucose conditions [12].
Phagocytosis Quantification: BV-2 cells were incubated with pH-sensitive fluorescent E. coli particles, and phagocytic capacity was measured via flow cytometry based internalization of fluorescent particles [12].
Migration Capacity: Evaluated using transwell migration assays, measuring the ability of microglial cells to migrate toward chemoattractant signals [12].
Cytokine Secretion Profiling: Multiplex ELISA arrays were employed to quantify secretion levels of pro-inflammatory cytokines (IL-6, TNF, IL-17, IL-12p70) from microglial cell supernatants under various conditions [72] [12].
Signaling Pathways and Molecular Mechanisms
Diagram 1: NDR2 Signaling Pathway in Microglial Metabolic Adaptation. This diagram illustrates NDR2's central role in regulating microglial responses to high-glucose stress, integrating metabolic adaptation with inflammatory output.
The molecular mechanism of NDR2 action involves its unique peroxisomal localization, enabled by the C-terminal GKL sequence that serves as a peroxisomal targeting signal [4]. This specific subcellular positioning allows NDR2 to interface with metabolic processes and influence cytoskeletal dynamics through phosphorylation of downstream targets including Rabin8, which subsequently activates Rab8 GTPase involved in vesicular trafficking [4]. Under high-glucose conditions, NDR2 upregulation enables metabolic flexibility by enhancing mitochondrial respiration capacity, while simultaneously coordinating phagocytic and migratory functions through cytoskeletal reorganization [72] [12]. When NDR2 function is compromised, microglial cells exhibit impaired metabolic adaptation alongside elevated pro-inflammatory cytokine secretion, creating a neuroinflammatory environment that contributes to diabetic retinopathy progression [72].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagents for Investigating NDR2 Function in Microglial Cells
| Reagent/Cell Line | Specific Application | Function/Utility |
|---|---|---|
| BV-2 Mouse Microglial Cells | In vitro modeling of microglial function [72] [12] | Immortalized microglial cell line retaining key microglial properties |
| CRISPR-Cas9 Ndr2 sgRNA | Targeted NDR2 downregulation [12] | Specific genomic editing to investigate NDR2 loss-of-function |
| Anti-NDR2 Antibody (#STJ94368) | Immunodetection of NDR2 protein [12] | Targets C-terminal region (aa 380-460) for Western blot/ICC |
| Anti-NDR1/2 Antibody (#sc-271703) | Simultaneous NDR1/2 detection [12] | Targets N-terminal region (aa 1-100) recognizing both kinases |
| Anti-IBA1 Antibody | Microglial identification [12] | Specific marker for microglial cells and macrophages |
| Seahorse Analyzer | Metabolic function assessment [12] | Measures mitochondrial respiration and glycolytic function |
| pHrodo E. coli Bioparticles | Phagocytosis quantification [12] | Fluorescent particles for tracking phagocytic activity |
The investigation of NDR2 in microglial adaptation to high-glucose environments reveals a critical regulatory node at the intersection of metabolic regulation and inflammatory response. The distinctive peroxisomal localization of NDR2, contrasted with NDR1's nuclear distribution, underscores the functional specialization between these highly similar kinases [4] [6]. In the context of diabetic retinopathy, NDR2 emerges as a promising therapeutic target whose modulation could potentially mitigate microglial-driven neuroinflammation while preserving beneficial homeostatic functions [72] [12]. Future research should explore the specific substrates through which NDR2 coordinates metabolic and inflammatory pathways and investigate whether pharmacological manipulation of NDR2 activity can restore microglial homeostasis in diabetic conditions, potentially offering new avenues for preventing vision loss in diabetic patients.
The Nuclear Dbf2-related (NDR) kinases NDR1 (STK38) and NDR2 (STK38L) are serine/threonine kinases belonging to the NDR/LATS subfamily of the Hippo signaling pathway. Despite sharing 87% amino acid sequence identity, they exhibit distinct subcellular localization: NDR1 is primarily nuclear, while NDR2 displays a punctate cytoplasmic distribution [3] [4] [6]. This fundamental difference in localization is conferred by a C-terminal peroxisome-targeting signal type 1 (PTS1) motif (Gly-Lys-Leu) in NDR2, which facilitates its binding to the Pex5p receptor and subsequent import into peroxisomes—a mechanism absent in NDR1, which terminates in Ala-Lys [4]. This comparative guide synthesizes current research to objectively evaluate how these distinct localization patterns dictate specialized functions for NDR1 and NDR2 in cancer, retinopathy, and infection models, providing essential experimental data and methodologies for researchers and drug development professionals.
Subcellular Localization: Mechanisms and Functional Consequences
The distinct subcellular distributions of NDR1 and NDR2 are a primary determinant of their non-overlapping functions. The table below summarizes the key localization mechanisms and their direct functional implications.
Table 1: Mechanisms and Functional Consequences of NDR1 and NDR2 Localization
| Kinase | Primary Localization | Targeting Motif/Mechanism | Key Functional Consequence | Validated Experimental Evidence |
|---|---|---|---|---|
| NDR1 | Nuclear [3] [6] | Lacks PTS1; C-terminal Ala-Lys [4] | Regulation of transcriptional processes (e.g., miR146a) [3] | Immunofluorescence; subcellular fractionation [3] [6] |
| NDR2 | Cytoplasmic (Punctate) / Peroxisomal [4] [6] | C-terminal PTS1 (Gly-Lys-Leu) binds Pex5p receptor [4] | Promotion of primary cilium formation [4] | Co-localization with catalase/CFP-SKL; Pex5p co-immunoprecipitation; rescue with NDR2(ΔL) mutant [4] |
Experimental Protocol: Validating Peroxisomal Localization of NDR2
The following methodology, adapted from Chiba et al. [4], is critical for confirming NDR2's unique localization.
- Cell Culture and Transfection: Culture appropriate cell lines (e.g., RPE1 or HeLa). Transfect with plasmids encoding fluorescently tagged proteins: YFP-NDR2, YFP-NDR1 (control), and CFP-SKL (a peroxisomal marker).
- Immunofluorescence Staining:
- Fix cells 24-48 hours post-transfection.
- Permeabilize and block cells using standard protocols.
- Incubate with primary antibodies against organelle markers (e.g., catalase for peroxisomes, GM130 for Golgi, LAMP1 for lysosomes).
- Incubate with fluorescently conjugated secondary antibodies.
- Image Acquisition and Analysis: Acquire high-resolution confocal microscopy images. Analyze for co-localization of YFP-NDR2 fluorescence with the peroxisomal marker (CFP-SKL or anti-catalase) using Pearson's correlation coefficient. NDR1 should serve as a negative control.
- Biochemical Validation (Subcellular Fractionation):
- Lyse YFP-NDR2-expressing cells and prepare a post-nuclear supernatant (PNS).
- Fractionate the PNS using iodixanol density gradient ultracentrifugation.
- Analyze fractions by Western blotting for YFP-NDR2 and the peroxisomal protein Pex14p. Co-sedimentation confirms peroxisomal localization.
- Functional Rescue: To prove the functional importance of localization, repeat ciliogenesis assays (e.g., in NDR2-knockdown cells) by reconstituting with wild-type NDR2 versus the peroxisome-targeting deficient mutant NDR2(ΔL).
Diagram 1: NDR2 Peroxisomal Targeting Pathway. The C-terminal GKL motif of NDR2 binds the Pex5p receptor, facilitating its import into peroxisomes, a step crucial for its role in promoting ciliogenesis.
Functional Comparisons in Disease Models
The differential localization of NDR1 and NDR2 translates into specialized and often opposing roles across pathological contexts.
Cancer
NDR kinases play complex, context-dependent roles in carcinogenesis, with NDR2 emerging as a prominent oncogene in several cancers.
Table 2: Comparative Roles of NDR1 and NDR2 in Cancer Models
| Disease Context | NDR1 Findings | NDR2 Findings | Key Experimental Data |
|---|---|---|---|
| Lung Cancer | Interactome analysis suggests distinct partners from NDR2 [10]. | Promotes proliferation, migration, invasion; regulates vesicle trafficking and autophagy [10]. | Proteomic comparison of NDR1 vs. NDR2 interactomes in HBEC-3, H2030, and H2030-BrM3 cells [10]. |
| Intestinal Epithelium | Dual ablation promotes colon carcinogenesis [4]. | Dual ablation promotes colon carcinogenesis [4]. | Ndr1/2 knockout mouse models show YAP dysregulation and tumor formation [4]. |
| Lymphoma | Ndr1 KO mice predisposed to T-cell lymphoma [4]. | Not strongly implicated [4]. | Genetic knockout mouse models [4]. |
Retinopathy and Neurodegeneration
Both kinases are critical for neuronal health, but NDR2 has a more defined role in specific retinopathies.
Table 3: Comparative Roles of NDR1 and NDR2 in Retinopathy and Neurodegeneration
| Disease Context | NDR1 Findings | NDR2 Findings | Key Experimental Data |
|---|---|---|---|
| Diabetic Retinopathy | Limited available data. | Upregulated in microglia under high glucose; regulates metabolic adaptation, phagocytosis, and migration; knockout increases pro-inflammatory cytokines (IL-6, TNF, IL-17) [12]. | CRISPR-Cas9 partial KO in BV-2 microglial cells; Seahorse analyzer for metabolism; ELISA for cytokines [12]. |
| Early Retinal Degeneration | Single KO shows minor retinal phenotypes [38]. | Causal gene mutation in canine early retinal degeneration (erd); crucial for photoreceptor and amacrine cell homeostasis [38]. | Ndr2 KO mouse models show aberrant proliferation of amacrine cells and rod opsin mislocalization [38]. |
| General Neurodegeneration | Single neuronal KO has minimal impact [73]. | Single neuronal KO has minimal impact [73]. | Dual knockout in excitatory neurons causes cortical/hippocampal neurodegeneration, impaired endocytosis, and autophagy defects [73]. |
Infection and Inflammation
NDR1 and NDR2 play distinct, sometimes opposing, roles in regulating innate immune responses.
Table 4: Comparative Roles of NDR1 and NDR2 in Infection and Inflammation
| Immune Context | NDR1 Function | NDR2 Function | Key Experimental Data |
|---|---|---|---|
| TLR9 Signaling (Bacterial CpG DNA) | Negative regulator. Binds Smurf1 to degrade MEKK2, inhibiting ERK1/2 and cytokine production (TNF-α, IL-6) [3]. | Negative regulator (similar to NDR1). Knockdown increases CpG-induced IL-6 [3]. | Stk38-deficient mice show higher cytokine levels and mortality after E. coli infection or sepsis [3]. |
| RIG-I Signaling (Antiviral) | Positive regulator. Binds miR146a intergenic region to promote STAT1 translation and type I IFN production [3]. | Positive regulator. Directly associates with RIG-I/TRIM25 to enhance RIG-I ubiquitination and type I IFN production [3]. | Knockdown/knockout studies with viral infection models (e.g., poly I:C stimulation) [3]. |
Diagram 2: NDR Kinases in Innate Immunity. NDR1/2 act as negative regulators of TLR9-mediated inflammation. In contrast, they are positive regulators of RIG-I-mediated antiviral immunity, albeit through distinct mechanisms.
The Scientist's Toolkit: Key Research Reagents and Models
The following table compiles essential reagents and models used in NDR1/2 research, as derived from the cited experimental data.
Table 5: Essential Research Reagents and Models for NDR1/2 Investigation
| Reagent / Model | Specification / Example | Primary Function in Research | Key Citation |
|---|---|---|---|
| Knockout Mouse Models | Ndr1 constitutive KO; Ndr2-floxed (crossed with NEX-Cre for neuronal KO) | Studying systemic and tissue-specific physiological functions and compensation in vivo. | [73] [38] |
| CRISPR-Cas9 Systems | All-in-one plasmid with sgRNA targeting exon 7 of Ndr2/Stk38l in BV-2 cells | Creating partial or complete gene knockdown/knockout in cell lines for functional studies. | [12] |
| Cell Lines | BV-2 (microglia), RPE1 (retinal pigment epithelium), HeLa, HEK293 | Conducting in vitro assays for signaling, localization, metabolism, and proliferation. | [4] [12] |
| Plasmid Constructs | YFP/CFP-tagged NDR1/2; NDR2(ΔL) mutant; CFP-SKL (peroxisome marker) | Visualizing subcellular localization and conducting structure-function studies. | [4] |
| Specific Antibodies | Anti-NDR1/2 (N-terminus); Anti-NDR2 (C-terminus, aa 380-460); Anti-IBA1 (microglia) | Detecting protein expression, localization (immunocytochemistry), and levels (Western blot). | [12] [38] |
| Metabolic Assay Kits | Seahorse XF Analyzer Kits (e.g., Mito Stress Test) | Profiling mitochondrial respiration and glycolytic function in live cells. | [12] |
The distinct subcellular destinies of NDR1 and NDR2, dictated by fundamental differences in their C-terminal sequences, establish a paradigm for functional specialization within a highly conserved kinase family. NDR1's nuclear presence correlates with roles in transcriptional regulation, while NDR2's peroxisomal and cytoplasmic localization underlies its unique capacity to govern ciliogenesis, vesicle trafficking, and metabolic adaptation. In disease, this divergence is critical: NDR2 has emerged as a central player in retinopathies and as an oncogene in lung cancer, whereas NDR1 appears more involved in lymphomagenesis. Their roles in infection are nuanced, jointly suppressing TLR9-driven inflammation but promoting RIG-I-mediated antiviral defense. Future therapeutic strategies targeting these kinases must, therefore, account for their distinct localization mechanisms and context-dependent functions. Research efforts should prioritize the full elucidation of the NDR1 and NDR2 interactomes and the development of highly specific inhibitors to exploit their differential roles in human disease.
Conclusion
The distinct subcellular localization of NDR1 and NDR2 is not a mere curiosity but a fundamental biological mechanism that enables these homologous kinases to govern largely separate cellular domains and functions. NDR1 operates as a nuclear regulator influencing gene expression and potentially DNA damage responses, while cytoplasmic NDR2 directly controls cytoskeletal dynamics, vesicle trafficking, and membrane receptor signaling. This functional specialization has direct implications for human disease, positioning NDR2 as a key player in neuroinflammatory conditions like diabetic retinopathy and integrin-mediated synaptic deficits, and NDR1 as a modulator of antiviral responses and inflammatory cytokine production. Future research must leverage the methodological frameworks outlined here to identify unique downstream substrates for each kinase and elucidate the potential for cross-talk. For drug development, these kinases present promising, distinct targets; inhibiting cytoplasmic NDR2 could ameliorate microglia-driven inflammation in retinopathy, while modulating nuclear NDR1 might fine-tune antiviral immunity. The continued dissection of the NDR1/2 localization-function paradigm will undoubtedly yield novel insights and therapeutic opportunities across immunology, neuroscience, and oncology.
References
- 1. Human Mob proteins regulate the NDR1 and NDR2 serine- ... [https://pubmed.ncbi.nlm.nih.gov/15067004/]
- 2. Chemical genetic identification of NDR1/2 kinase substrates ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3333840/]
- 3. The Emerging Roles of NDR1/2 in Infection and Inflammation [https://pmc.ncbi.nlm.nih.gov/articles/PMC7105721/]
- 4. Localization of Protein Kinase NDR2 to Peroxisomes and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5354478/]
- 5. The NDR/LATS protein kinases in neurobiology [https://www.sciencedirect.com/science/article/pii/S0171933523000481]
- 6. Human Mob Proteins Regulate the NDR1 and NDR2 ... [https://www.sciencedirect.com/science/article/pii/S0021925820665796]
- 7. Localization of Protein Kinase NDR2 to Peroxisomes and ... [https://pubmed.ncbi.nlm.nih.gov/28122914/]
- 8. Human NDR Kinases Are Rapidly Activated by MOB Proteins ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1234321/]
- 9. Mechanism of Activation of NDR (Nuclear Dbf2-related) ... [https://www.sciencedirect.com/science/article/pii/S0021925820730991]
- 10. NDR2 kinase: A review of its physiological role and ... [https://www.sciencedirect.com/science/article/pii/S0141813025042084]
- 11. NDR1/2's Role in Infection [https://www.news-medical.net/life-sciences/NDR12e28099s-Role-in-Infection.aspx]
- 12. NDR2 Kinase Regulates Microglial Metabolic Adaptation and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12609617/]
- 13. Hypoxia-induced activation of NDR2 underlies brain ... [https://www.nature.com/articles/s41419-023-06345-3]
- 14. The Emerging Roles of NDR1/2 in Infection and Inflammation [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2020.00534/full]
- 15. Identification of N-Terminal Lobe Motifs that Determine the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2768531/]
- 16. Structural Basis for Auto-Inhibition of the NDR1 Kinase ... [https://www.sciencedirect.com/science/article/pii/S0969212618301783]
- 17. Regulation of NDR Protein Kinase by Hydrophobic Motif ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1316964/]
- 18. mediated regulation of NDR protein kinase through ... [https://www.sciencedirect.com/science/article/pii/S0021925819325918]
- 19. Constitutively active NDR1-PIF kinase functions independent ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4049220/]
- 20. Regulation of NDR1 activity by PLK1 ensures proper ... [https://www.nature.com/articles/srep10449]
- 21. Monopolar spindle-one-binder protein 2 regulates the ... [https://www.spandidos-publications.com/10.3892/ol.2018.7952]
- 22. Mechanism of activation of NDR (nuclear Dbf2-related) ... [https://pubmed.ncbi.nlm.nih.gov/15197186/]
- 23. The NDR family of kinases: essential regulators of aging - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11129689/]
- 24. A review of nuclear Dbf2-related kinase 1 (NDR1) protein ... [https://www.sciencedirect.com/science/article/abs/pii/S0141813023060877]
- 25. Discovery of a small-molecule NDR1 agonist for prostate ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2024.1367358/full]
- 26. NDR2 promotes the antiviral immune response via facilitating ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6365120/]
- 27. Human NDR Kinases Control G1/S Cell Cycle Transition ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3135299/]
- 28. Loss of NDR1/2 kinases impairs endomembrane trafficking ... [https://www.life-science-alliance.org/content/6/2/e202201712]
- 29. Guide to Antibody Validation Techniques [https://www.neobiotechnologies.com/resources/antibody-validation-methods/?srsltid=AfmBOoqBD_BTysGeSvgxdYD-JWCzdr_Ob035pC36i380PeYo_F2K4yxS]
- 30. Antibody validation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3891910/]
- 31. Visualizing cell structure and function with point-localization ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4221848/]
- 32. Identification of the reporter gene combination that shows high ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0297273]
- 33. CRISPR/Cas9-mediated SHP-1-knockout T cells combined ... [https://www.sciencedirect.com/science/article/pii/S2589004225005279]
- 34. Assessment of RNA interference effectiveness mediated by ... [https://www.sciencedirect.com/science/article/abs/pii/S0965174825001626]
- 35. Systematic analysis of siRNA and mRNA features impacting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12205987/]
- 36. CRISPR-StAR enables high-resolution genetic screening ... [https://www.nature.com/articles/s41587-024-02512-9]
- 37. Drug and siRNA screens identify ROCK2 as a therapeutic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12009310/]
- 38. Ndr kinases regulate retinal interneuron proliferation and ... [https://www.nature.com/articles/s41598-018-30492-9]
- 39. Fractionation of Cells - Molecular Biology of the Cell - NCBI [https://www.ncbi.nlm.nih.gov/books/NBK26936/]
- 40. Subcellular Fractionation: Reliable Protocols Explained [https://bitesizebio.com/20047/subcellular-fractionation-protocol-explained/]
- 41. Subcellular fractionation protocol [https://www.abcam.com/en-us/technical-resources/protocols/subcellular-fractionation]
- 42. Overview of Cell Fractionation and Organelle Isolation [https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/cell-fractionation-organelle-isolation.html]
- 43. The Roles of NDR Protein Kinases in Hippo Signalling - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4880841/]
- 44. NDR1 modulates the UV-induced DNA-damage ... [https://pubmed.ncbi.nlm.nih.gov/25912875/]
- 45. Loss of NDR1/2 kinases impairs endomembrane trafficking ... [https://pubmed.ncbi.nlm.nih.gov/36446521/]
- 46. The NDR family of kinases: essential regulators of aging [https://www.frontiersin.org/journals/molecular-neuroscience/articles/10.3389/fnmol.2024.1371086/full]
- 47. The Serine/Threonine Kinase Ndr2 Controls Integrin ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6609001/]
- 48. NDR2 Kinase Regulates Microglial Metabolic Adaptation ... [https://www.mdpi.com/1422-0067/26/21/10630/review_report]
- 49. Modeling the neuroimmune system in Alzheimer's and ... [https://jneuroinflammation.biomedcentral.com/articles/10.1186/s12974-024-03024-8]
- 50. NDR Kinases Are Essential for Somitogenesis and Cardiac ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0136566]
- 51. NDR Functions as a Physiological YAP1 Kinase in the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4426889/]
- 52. Functional analysis of AKT1 knockout in fibrosarcoma cells ... [https://www.nature.com/articles/s10038-025-01436-9]
- 53. A strategy to disentangle direct and indirect effects on (de) ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10882499/]
- 54. Genetic algorithm as an optimization tool for the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6407725/]
- 55. Genetic Optimization in Uncovering Biologically ... [https://www.mdpi.com/2673-7426/4/1/45]
- 56. NDR2 promotes the antiviral immune response via facilitating TRIM25-mediated RIG-I activation in macrophages - PubMed [https://pubmed.ncbi.nlm.nih.gov/30775439/]
- 57. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2737813/]
- 58. Molecular mechanism of influenza A NS1-mediated ... [https://www.nature.com/articles/s41467-018-04214-8]
- 59. A RIG-I–like receptor directs antiviral responses to ... [https://www.sciencedirect.com/science/article/pii/S0021925817489848]
- 60. The Serine/Threonine Kinase NDR2 Regulates Integrin ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12120816/]
- 61. Ndr2 Kinase Controls Neurite Outgrowth and Dendritic ... [https://www.frontiersin.org/journals/molecular-neuroscience/articles/10.3389/fnmol.2018.00066/full]
- 62. The NDR/LATS protein kinases in neurobiology: Key regulators of cell... [https://pubmed.ncbi.nlm.nih.gov/37327741/]
- 63. The roles of NDR /2 kinases in 1 membrane... - UCL Discovery neuronal [https://discovery.ucl.ac.uk/id/eprint/10143347/]
- 64. The Hippo signalling pathway and its implications in ... [https://www.nature.com/articles/s41392-022-01191-9]
- 65. The components and regulation of the Hippo pathway and its ... [https://cancerci.biomedcentral.com/articles/10.1186/s12935-025-03946-0]
- 66. The multifaceted role of YAP in the tumor ... [https://www.nature.com/articles/s12276-025-01551-9]
- 67. Transcriptional Regulation of the Hippo Pathway [https://www.mdpi.com/2073-4409/11/14/2225]
- 68. Targeting the Hippo/YAP Pathway: A Promising Approach for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12394893/]
- 69. Hippo/YAP signaling pathway in colorectal cancer [https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2025.1545952/full]
- 70. Endogenous YAP/TAZ partitioning in TEAD condensates ... [https://www.sciencedirect.com/science/article/abs/pii/S1097276525006641]
- 71. The Regulation of the Hippo Signalling Pathway Effector YAP ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12607357/]
- 72. NDR2 Kinase Regulates Microglial Metabolic Adaptation ... [https://pubmed.ncbi.nlm.nih.gov/41226665/]
- 73. Loss of NDR1/2 kinases impairs endomembrane trafficking ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9711861/]