MOB1 vs MOB2: Decoding Binding Specificity and Functional Regulation of NDR Kinases in Hippo Signaling
This article provides a comprehensive analysis of the distinct binding specificities of MOB1 and MOB2 coactivators for NDR/LATS kinases, crucial regulators in Hippo signaling pathways.
MOB1 vs MOB2: Decoding Binding Specificity and Functional Regulation of NDR Kinases in Hippo Signaling
Abstract
This article provides a comprehensive analysis of the distinct binding specificities of MOB1 and MOB2 coactivators for NDR/LATS kinases, crucial regulators in Hippo signaling pathways. We explore the structural basis for selective kinase-coactivator complex formation, examining how MOB1 activates LATS kinases while MOB2 exhibits a context-dependent regulatory relationship with NDR kinases. The content covers experimental methodologies for studying these interactions, addresses common research challenges, and validates functional consequences in physiological and disease contexts, particularly cancer. This synthesis aims to equip researchers and drug development professionals with mechanistic insights and practical frameworks for targeting these specific protein interactions in therapeutic development.
Structural Foundations and Evolutionary Conservation of MOB-NDR Kinase Interactions
The Hippo signaling pathway is an evolutionarily conserved system crucial for controlling cell proliferation, morphogenesis, and organ size in eukaryotes. At the heart of this pathway are the NDR/LATS kinases (Nuclear Dbf2-related / Large Tumor Suppressor kinases), which belong to the AGC family of serine-threonine protein kinases. These kinases form functional complexes with MOB coactivator proteins (Monopolar spindle one-binder), an association that is essential for kinase activity and pathway function [1] [2].
The NDR/LATS family is divided into two main subfamilies: the LATS kinases (including Dbf2 and Dbf20 in yeast, LATS1/2 in mammals) that primarily associate with MOB1 proteins, and the NDR kinases (including Cbk1 in yeast, NDR1/STK38 and NDR2/STK38L in mammals) that specifically bind MOB2 proteins [1] [3]. This specific kinase-cofactor pairing is maintained from yeast to humans despite significant sequence conservation among MOB proteins, indicating strong evolutionary pressure to preserve binding specificity [1] [4].
Table 1: Core NDR/LATS Kinases and Their MOB Cofactors Across Model Organisms
| Organism | NDR-subfamily Kinases | MOB Cofactor | LATS-subfamily Kinases | MOB Cofactor | Biological Function |
|---|---|---|---|---|---|
| S. cerevisiae (Budding Yeast) | Cbk1 | Mob2 | Dbf2, Dbf20 | Mob1 | RAM network: cell separation, morphogenesis; MEN: mitotic exit, cytokinesis [1] [2] |
| D. melanogaster (Fruit Fly) | – | dMob2 | Warts | dMob1 (Mats) | Hippo pathway: tissue growth, neuromuscular junction morphology [4] [5] |
| H. sapiens (Human) | NDR1/STK38, NDR2/STK38L | MOB2 | LATS1, LATS2 | MOB1A/B | Cell cycle progression, DNA damage response, neuronal development [3] [4] [6] |
Structural Basis of Kinase-Cofactor Interactions
The association between NDR/LATS kinases and MOB cofactors is mediated through a characteristic N-terminal regulatory (NTR) region in the kinases that binds to a specific surface on the MOB proteins. Structural analyses of complexes such as Saccharomyces cerevisiae Cbk1NTR–Mob2 and Dbf2NTR–Mob1 have revealed that the NTR forms a V-shaped helical hairpin that docks onto the MOB protein [1]. This NTR–Mob interface serves as a common structural platform that mediates kinase–cofactor binding across the entire NDR/LATS family.
The MOB cofactor plays a critical role in kinase activation by organizing the NTR to interact with the AGC kinase C-terminal hydrophobic motif (HM), which is involved in allosteric regulation. This Mob-organized NTR appears to mediate association of the HM with an allosteric site on the N-terminal kinase lobe, facilitating kinase activation [1]. This mechanism represents a distinctive kinase regulation mechanism unique to NDR/LATS kinases.
Molecular Determinants of MOB1 vs. MOB2 Specificity
The specific pairing of LATS kinases with MOB1 and NDR kinases with MOB2 is enforced by discrete structural elements rather than broadly distributed interface differences. Several key determinants have been identified:
- Short specificity motifs: A short motif in the Mob structure that differs between Mob1 and Mob2 strongly contributes to molecular recognition [1]
- Key residue interactions: Alteration of specific residues in the Cbk1 NTR allows association with the noncognate Mob cofactor, demonstrating that specificity is restricted to discrete sites [1]
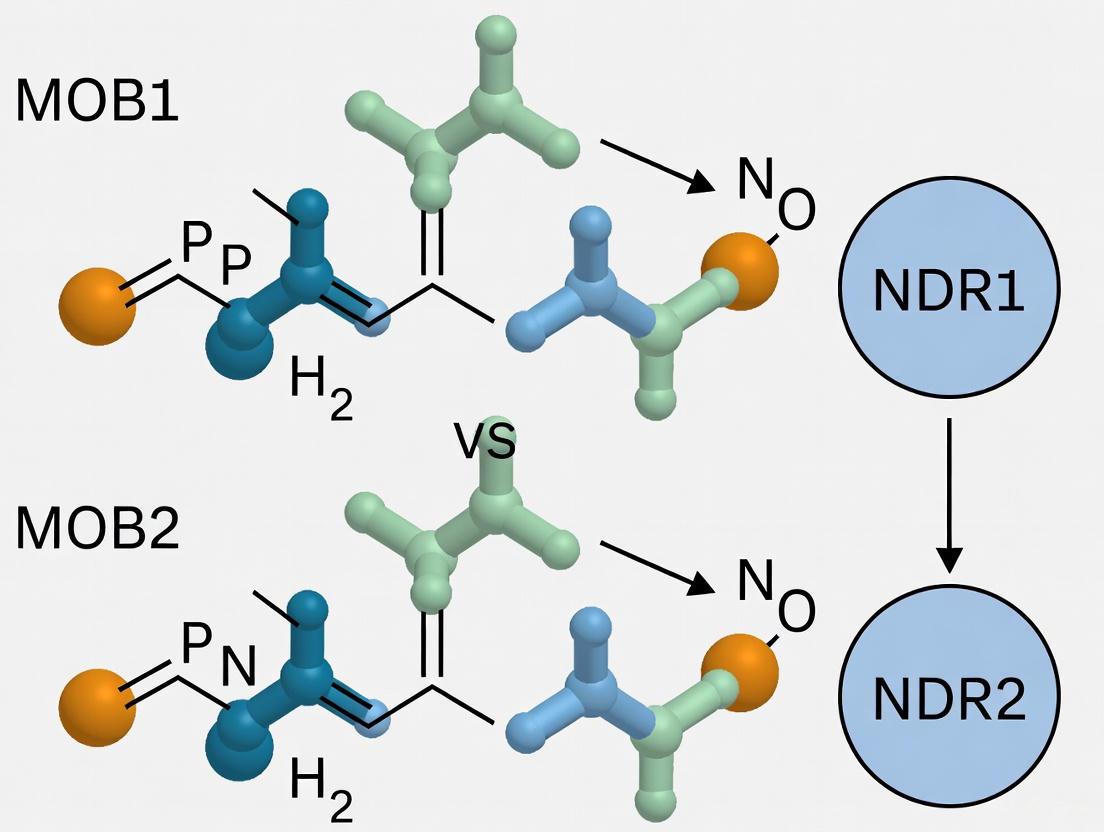
- Competitive binding mechanisms: In mammalian cells, MOB2 competes with MOB1 for NDR binding, with MOB1/NDR complexes associated with increased NDR kinase activity while MOB2/NDR complexes show diminished activity [4]
Table 2: Structural and Functional Differences Between MOB1 and MOB2 Complexes
| Characteristic | MOB1 Complexes | MOB2 Complexes |
|---|---|---|
| Kinase Partners | LATS kinases (Dbf2/Dbf20 in yeast, LATS1/2 in mammals) [1] | NDR kinases (Cbk1 in yeast, NDR1/2 in mammals) [1] [3] |
| Cellular Functions | Mitotic exit, cytokinesis, Hippo pathway regulation, YAP/TAZ inhibition [2] | Cell morphogenesis, polarization, DNA damage response, neural development [4] [6] |
| Subcellular Localization | Cytoplasmic, spindle pole bodies, cell cortex [2] | Nuclear and cytoplasmic, punctate cytoplasmic distribution [3] |
| Activation Outcome | Enhanced kinase activity toward specific substrates (e.g., YAP/TAZ) [2] | Varied effects: can stimulate or diminish kinase activity depending on context [4] |
| Regulatory Role | Core pathway component [2] | Context-dependent modulator [4] |
Functional Consequences of Kinase-Cofactor Interactions
Activation Mechanisms and Downstream Signaling
The binding of MOB cofactors to NDR/LATS kinases dramatically influences kinase activity and substrate specificity. For mammalian NDR kinases, association with MOB2 strongly stimulates catalytic activity [3]. This activation depends on the phosphorylation status of key regulatory sites, including the hydrophobic motif (HM) and activation loop (AL). The MOB-organized NTR facilitates proper positioning of the phosphorylated HM, enabling optimal orientation of the kinase αC helix, a component critical for kinase activation [1].
The functional outcomes of NDR/LATS-MOB complexes differ significantly between the two subfamilies:
- MOB1-LATS complexes: Primarily regulate cell proliferation and organ size through phosphorylation of YAP/TAZ transcriptional coactivators, leading to their cytoplasmic sequestration and degradation [2]
- MOB2-NDR complexes: Govern diverse processes including cell morphogenesis, polarization, DNA damage response, and neuronal development through phosphorylation of distinct substrates [4] [6]
Biological Functions in Cellular Processes
MOB2-NDR kinase signaling plays critical roles in several fundamental cellular processes:
- Cell cycle and DNA damage response: Endogenous MOB2 is required to prevent accumulation of DNA damage and avoid undesired activation of cell cycle checkpoints. MOB2 depletion triggers p53/p21-dependent G1/S cell cycle arrest [4]
- Neural development: NDR kinases regulate vesicle trafficking, polarity, and morphogenesis in neuronal tissues. Deletion of Ndr kinases leads to concurrent apoptosis and proliferation of retinal neurons [6]
- Cell proliferation control: MOB2 competes with MOB1 for NDR binding, potentially creating a regulatory switch that modulates NDR kinase activity in response to cellular cues [4]
Experimental Approaches for Studying MOB-Kinase Interactions
Structural Biology Methodologies
Protein Expression and Purification
- Challenge: Recombinant expression of monomeric Mob2 in E. coli can be problematic due to stability issues
- Solution: Engineering of zinc-binding Mob2 (V148C Y153C) that recapitulates zinc-binding motifs found in metazoan Mob2 orthologs, enabling suitable E. coli expression for biochemistry [1]
- Protocol: Express NTR regions (e.g., Cbk1NTR residues 251-351) and Mob proteins in E. coli, purify using affinity chromatography, and form complexes for crystallography
Crystallographic Data Collection and Structure Determination
- Crystallization: Co-crystallize NTR-Mob complexes using vapor diffusion methods
- Data Collection: Collect X-ray diffraction data at synchrotron sources (e.g., wavelength ~0.978-1.000 Å)
- Structure Determination: Solve structures using molecular replacement with existing related structures as search models
- Refinement: Iterative model building and refinement to achieve final structures with R-work/R-free values typically around 0.23/0.30 [1]
Table 3: Crystallographic Data Collection and Refinement Statistics for Representative NTR-MOB Complexes
| Parameter | Dbf2NTR–Mob1 | Cbk1NTR–Mob2 | Cbk1–Mob2–pepSsd1 |
|---|---|---|---|
| Resolution (Å) | 46.9–3.5 (3.59–3.50) | 39.9–2.8 (2.87–2.80) | 44.60–3.15 (3.23–3.15) |
| Space Group | P61 2 2 | P41 2 12 | C1 2 1 |
| Cell Dimensions (Å) | a=108.28, b=108.28, c=134.76 | a=126.23, b=126.27, c=49.34 | a=138.43, b=79.99, c=117.59 |
| Completeness (%) | 99.9 (100) | 99.8 (100) | 99.7 (99.7) |
| R-work/R-free | 0.2292/0.2631 | 0.2490/0.2838 | 0.2310/0.2983 |
| Ramachandran Favored (%) | 93.5 | 93 | 76 |
| PDB Reference | Not specified | Not specified | Not specified |
Interaction Mapping Techniques
Proximity-Dependent Biotin Identification (BioID)
- Principle: Fuse MOB proteins to a promiscuous biotin ligase (BirA*) that biotinylates proximal proteins within ~10 nm
- Cell Systems: Generate tetracycline-inducible HEK293 and HeLa Flp-In T-REx cells expressing BirA*-FLAG-MOB fusions
- Controls: Include BirA-FLAG and BirA-FLAG-EGFP as negative controls
- Validation: Confirm interactions by affinity purification-mass spectrometry and functional assays [7]
Binding Affinity and Specificity Assays
- Quantitative approaches: Fluorescence polarization (FP) to measure binding affinities of wild-type and mutant complexes
- Specificity mapping: Systematic mutagenesis of interface residues to identify determinants of MOB1 vs. MOB2 specificity [1]
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagents for Studying NDR/LATS-MOB Interactions
| Reagent/Solution | Function/Application | Examples/Specifications |
|---|---|---|
| Zinc-binding Mob2 mutant | Stabilizes Mob2 for structural studies | Mob2 V148C Y153C engineering recapitulates metazoan zinc-binding motif [1] |
| NTR construct plasmids | Expression of kinase regulatory regions | Cbk1NTR (251-351), Dbf2NTR for binding studies [1] |
| Tetracycline-inducible BioID cell lines | Proximity-dependent interaction mapping | HEK293 and HeLa Flp-In T-REx BirA*-FLAG-MOB cells [7] |
| Crystallization screens | Optimization of protein crystal growth | Commercial sparse matrix screens for various space groups [1] |
| Phospho-specific antibodies | Detection of HM and AL phosphorylation | Critical for monitoring kinase activation status [1] |
| Docking motif peptides | Substrate interaction studies | pepSsd1 for Cbk1 substrate docking characterization [1] |
| Kinase activity assays | Measuring NDR/LATS kinase activation | Radioactive or luminescent kinase assays with specific substrates [3] |
Future Perspectives and Research Directions
The study of NDR/LATS kinases and their MOB cofactors continues to evolve with several emerging research directions:
- MOB3 subfamily characterization: MOB3 proteins represent a poorly characterized branch that associates with the pro-apoptotic kinase MST1 rather than NDR/LATS kinases [4]. Recent BioID studies revealed an unexpected connection between MOB3C and the RNase P complex, suggesting potential roles in RNA biology [7]
- Therapeutic targeting: The distinct roles of NDR1 and NDR2 in processes like vesicle trafficking, autophagy, and immune response position them as potential therapeutic targets, particularly in cancer contexts [8] [6]
- Neurobiological functions: Emerging evidence indicates crucial roles for NDR kinases in neuronal development, homeostasis, and inflammation, suggesting potential therapeutic applications for neuronal diseases [6]
Understanding the precise molecular determinants governing MOB1 versus MOB2 binding specificity remains a crucial area of investigation, with implications for targeted therapeutic intervention in cancer and other diseases where Hippo signaling is disrupted.
The NDR/LATS family of kinases and their MOB coactivators constitute an evolutionarily conserved signaling module central to eukaryotic cell proliferation, morphogenesis, and survival. This whitepaper delineates the stringent binding specificity that defines the CBK1-Mob2 and DBF2-Mob1 paradigms, drawing on structural, biochemical, and functional evidence from yeast to human homologs. We examine the molecular determinants that enforce selective kinase-coactivator pairing, the downstream physiological consequences of these specific complexes, and their implications for therapeutic targeting. Within the broader context of MOB1 versus MOB2 binding specificity for NDR kinase research, this analysis underscores how conserved structural frameworks yield diverse, context-dependent biological outputs, from regulating mitotic exit to controlling organ size.
The NDR (Nuclear Dbf2-related) / LATS (Large Tumor Suppressor) family of AGC kinases represents a deeply conserved branch of eukaryotic signaling pathways [9]. These kinases, which include Saccharomyces cerevisiae Cbk1 and Dbf2, and their human counterparts NDR1/2 and LATS1/2, require binding to Mob (Mps one binder) coactivator proteins for their full activity and biological function [1] [3] [10]. A fundamental characteristic of this system is the high specificity of kinase-coactivator interactions: NDR-subfamily kinases (e.g., Cbk1) specifically associate with Mob2 proteins, while LATS-subfamily kinases (e.g., Dbf2) specifically associate with Mob1 proteins [1] [2]. This specificity is maintained despite significant sequence conservation within both the kinase and Mob protein families, suggesting strong evolutionary pressure to preserve discrete functional partnerships.
These specific kinase-Mob complexes form the core of ancient signaling networks, notably the Hippo pathway, which controls cell proliferation, morphogenesis, and apoptosis from yeast to humans [2] [9]. In budding yeast, the two primary networks are the Regulation of Ace2 and Morphogenesis (RAM) network, governed by Cbk1-Mob2, and the Mitotic Exit Network (MEN), governed by Dbf2/20-Mob1 [11] [2]. The functional segregation of these pathways underscores the critical importance of specific Mob pairing: despite simultaneous presence of all proteins in the cytosol, Cbk1–Mob1 or Dbf2–Mob2 complexes do not form, ensuring proper signaling fidelity [1].
Structural Basis of Kinase-MOB Specificity
Structural biology has been instrumental in elucidating the molecular basis for selective Mob recognition. The crystal structure of the Saccharomyces cerevisiae Cbk1 kinase bound to Mob2 provides a high-resolution model of an NDR kinase-Mob complex [1] [2]. The NDR/LATS kinases contain a distinctive N-terminal regulatory (NTR) region that directly binds the Mob coactivator. This NTR region forms a V-shaped helical hairpin that docks against a conserved surface on the Mob protein [1].
Table 1: Key Structural Features of Kinase-MOB Complexes
| Component | Structural Feature | Functional Role | Conservation |
|---|---|---|---|
| Kinase NTR | V-shaped helical hairpin | Mob coactivator binding | High from yeast to humans |
| Mob Protein | Conserved core fold | Kinase activation & localization | High from yeast to humans |
| Hydrophobic Motif (HM) | C-terminal motif | Allosteric kinase regulation | Conserved in AGC kinases |
| Specificity Motif | Variable loop in Mob | Determines Mob1 vs. Mob2 binding | Key residues differ between Mob1/2 |
The binding of Mob to the kinase NTR organizes this region, enabling it to interact with the kinase's C-terminal hydrophobic motif (HM), a critical element for allosteric kinase activation [1]. This Mob-organized NTR appears to mediate association of the HM with an allosteric site on the N-terminal kinase lobe, facilitating kinase activation. This mechanism represents a distinctive mode of kinase regulation unique to the NDR/LATS family.
Molecular Determinants of MOB1 vs. MOB2 Specificity
Specificity for cognate Mob binding is restricted by discrete molecular recognition sites rather than being broadly distributed across the interaction surface. Comparative structural analysis of Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes reveals that a short, variable motif within the Mob structure, which differs between Mob1 and Mob2, strongly contributes to molecular recognition [1]. Alteration of specific residues in this motif in Cbk1's NTR allows association with the non-cognate Mob cofactor, confirming that these discrete sites act as specificity gates.
The improved structural model of the Cbk1-Mob2 interface highlights the role of Mob binding in positioning the phosphorylated threonine (Thr-743 in Cbk1) within the kinase's HM. Upon Mob binding, this phosphorylated residue becomes proximal to a conserved arginine in the NTR, driving optimal positioning of the kinase's αC helix, a component critical for kinase activation [1]. This precise geometric arrangement is only achievable with the cognate Mob partner, thereby coupling binding specificity to catalytic activation.
Diagram 1: Structural basis of kinase-MOB specificity. The NTR and MOB specificity motif enforce selective binding, leading to stable, active cognate complexes.
Functional Consequences of Specific Pairing
The CBK1-Mob2 Complex in the RAM Network
The Cbk1-Mob2 complex is the central effector of the RAM network in budding yeast. This pathway is essential for controlling cell separation and polarized morphogenesis [11] [12]. During hyphal development in Candida albicans, Cbk1 and Mob2 localize to the tips of growing hyphae and the region of the septum, where they regulate the maintenance of polarisome components [11]. A key mechanism involves CDK-dependent phosphorylation of Mob2, which is essential for normal hyphal development. Mutations in CDK consensus sites within Mob2 significantly impair hyphal development, causing short hyphae with enlarged tips and illicit activation of cell separation programs [11].
A critical function of the Cbk1-Mob2 complex is the direct phosphorylation and regulation of the Ace2 transcription factor. Cbk1 phosphorylates Ace2 at specific sites matching its unusual consensus motif (strong preference for histidine at position -5: H-X-[K/R]-[K/R]-X-[S/T]), which controls Ace2's asymmetric localization to the daughter cell nucleus and its transcriptional activity [12]. This phosphorylation blocks Ace2's interaction with nuclear export machinery, trapping it in the daughter nucleus where it drives expression of genes required for cell separation [12].
The DBF2-Mob1 Complex in the Mitotic Exit Network
The Dbf2-Mob1 complex functions as a central component of the MEN, which controls cytokinesis and the transition from M phase to G1 [1] [2]. This pathway ensures the coordinated completion of mitosis and cell division. The Dbf2-Mob1 complex is recruited to the spindle pole bodies by the scaffold protein Nud1, where it becomes activated and promotes the release of the Cdc14 phosphatase from the nucleolus, leading to the reversal of mitotic CDK phosphorylation and mitotic exit [2].
The specific pairing of Dbf2 with Mob1, rather than Mob2, is essential for directing this complex to its correct mitotic functions. While the structural basis of activation is similar to Cbk1-Mob2, the downstream substrates and cellular localization differ dramatically, driven by the distinct protein-protein interaction networks enabled by Mob1 versus Mob2.
Conservation in Metazoans: From Flies to Humans
The functional specificity of Mob pairing is conserved in metazoans. Human NDR1 and NDR2 kinases associate specifically with MOB2 proteins, while LATS1 and LATS2 associate with MOB1 proteins [13] [3]. Human MOB proteins dramatically stimulate the catalytic activity of their cognate NDR kinases [3] [10]. The activation mechanism involves recruitment to cellular membranes, where membrane-targeted MOBs robustly promote NDR phosphorylation and activation within minutes of association with membranous structures [13].
In humans, the LATS1/2-MOB1 complex phosphorylates the YAP/TAZ transcriptional coactivators, leading to their cytoplasmic retention and degradation, thereby suppressing cell proliferation—a function disrupted in many cancers [2] [9]. The NDR1/2-MOB2 complex, while less characterized, contributes to neuronal morphogenesis, cell cycle progression, and control of the G1/S restriction point [9].
Table 2: Functional Specialization of Kinase-MOB Complexes Across Species
| Complex | Yeast Pathway | Primary Yeast Functions | Mammalian Homologs | Mammalian Functions |
|---|---|---|---|---|
| CBK1-Mob2 | RAM Network | Cell separation, polarized growth, Ace2 regulation | NDR1/2 - MOB2 | Neurite outgrowth, cell proliferation, Golgi organization |
| DBF2-Mob1 | MEN | Mitotic exit, cytokinesis, Cdc14 activation | LATS1/2 - MOB1 | Hippo signaling, YAP/TAZ phosphorylation, tumor suppression |
Experimental Analysis of Kinase-MOB Interactions
Key Methodologies and Reagents
Research into kinase-MOB specificity employs a multidisciplinary approach combining structural biology, biochemistry, and genetics. The following experimental protocols are central to this field.
Protocol 1: Structural Determination of Kinase-MOB Complexes
- Protein Expression and Purification: Express the kinase N-terminal regulatory region (NTR) and full-length Mob protein in E. coli. For unstable Mob proteins (e.g., wild-type Mob2), engineering a zinc-binding motif (e.g., Mob2 V148C Y153C) can improve stability and expression [1].
- Complex Formation: Mix purified kinase NTR and Mob proteins in equimolar ratios and purify the complex using size-exclusion chromatography.
- Crystallization: Screen for crystallization conditions using robotic systems. For Cbk1NTR–Mob2, crystals grew in condition containing 0.1 M sodium citrate tribasic dihydrate pH 5.5, 15% (v/v) 2-Propanol, 15% (w/v) PEG 4,000 [1].
- Data Collection and Structure Determination: Collect X-ray diffraction data (e.g., at 2.8 Å resolution for Cbk1NTR–Mob2). Solve the structure by molecular replacement using known Mob and kinase domain structures as search models. Refine the model iteratively [1] [2].
Protocol 2: Assessing MOB-Dependent Kinase Activation
- Kinase Assay Setup: Prepare reaction buffer containing 25 mM Tris-HCl pH 7.5, 5 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, 10 mM MgCl2.
- Mob Stimulation: Incubate NDR kinase with increasing concentrations of cognate or non-cognate Mob protein (e.g., 0-10 μM) for 15-30 minutes at 30°C.
- Phosphorylation Reaction: Initiate reaction by adding ATP mix (100 μM ATP containing [γ-32P]ATP). Use a suitable substrate such as the histone H1 or a specific peptide substrate (e.g., for Cbk1: a peptide derived from Ace2 containing the H-X-[K/R]-[K/R]-X-[S/T] motif) [12].
- Detection: Terminate reactions with SDS sample buffer, separate proteins by SDS-PAGE, and visualize phosphorylated substrates by autoradiography or phosphorimaging [3] [10].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Investigating Kinase-MOB Specificity
| Reagent/Solution | Function/Application | Example from Literature |
|---|---|---|
| Zinc-binding Mob2 Mutant | Stabilizes Mob2 for structural studies and biochemical assays | Mob2 V148C Y153C for E. coli expression [1] |
| λ-Phosphatase | Distinguishes phosphorylated protein isoforms; confirms phospho-mobility shifts on gels | Treatment of Mob2 extracts to confirm phosphorylation-dependent mobility shift [11] |
| Phospho-specific Antibodies | Detects activated, phosphorylated kinases; assesses kinase activation state | Antibodies against phosphorylated Ser281 and Thr444 of human NDR1 [13] |
| Membrane-Targeting Constructs | Investigates role of subcellular localization in kinase activation | Myristoylation/palmitylation motif fusions for membrane targeting of NDR/MOB [13] |
| Conditional Expression Systems | For inducible membrane translocation to study kinetics of activation | Chimeric hMOB with C1 domain for phorbol ester-induced membrane recruitment [13] |
| Peptide Scanning Arrays | Defines kinase phosphorylation consensus motifs | Used to determine Cbk1's unique basophilic motif with histidine at -5 [12] |
Visualization of Signaling Pathways and Molecular Interactions
Diagram 2: Specific kinase-MOB complexes in distinct signaling pathways. The CBK1-Mob2 and DBF2-Mob1 complexes function in separate pathways (RAM and MEN) with different inputs and biological outputs.
Discussion and Research Implications
The evolutionary conservation of specific CBK1-Mob2 and DBF2-Mob1 pairing from yeast to humans highlights the fundamental importance of this regulatory paradigm. The structural mechanisms enforcing this specificity are remarkably conserved, yet these core signaling modules have been adapted to control diverse biological processes across eukaryotes—from fungal cell polarity to mammalian organ size and neuronal development [2] [9].
From a therapeutic perspective, the NDR/LATS kinases and their Mob coactivators represent attractive targets, particularly in cancer where Hippo signaling is frequently disrupted. The discrete nature of the specificity determinants suggests it might be possible to develop small molecules that selectively disrupt specific kinase-Mob interactions or modulate the activity of particular complexes. Furthermore, the recent implication of NDR kinases in aging hallmarks—including cellular senescence, chronic inflammation, and loss of proteostasis—opens new avenues for research into therapeutic interventions for age-related diseases [9].
Future research directions should focus on: (1) elucidating the complete regulatory network of phosphorylation events controlling kinase-Mob complex assembly and activity; (2) developing selective chemical probes to manipulate specific kinase-Mob interactions in cellular and animal models; and (3) exploring the therapeutic potential of targeting these complexes in cancer, neurodegenerative diseases, and other age-associated conditions. The deep evolutionary conservation of these pathways from yeast to humans provides a powerful foundation for using model organisms to unravel fundamental mechanisms with direct relevance to human health and disease.
The NDR (Nuclear Dbf2-related) kinase family and their MOB (Mps one binder) coactivators constitute an evolutionarily conserved signaling module central to eukaryotic biology, governing processes from cell proliferation and morphogenesis to neuronal development and tumor suppression [14] [15]. The functional core of this module is the specific complex formed between the N-terminal Regulatory (NTR) domain of an NDR/LATS kinase and its cognate MOB protein. For researchers and drug development professionals, a precise understanding of this interface is paramount, as it dictates signaling specificity and presents a potential target for therapeutic intervention. This whitepaper delineates the structural principles governing NDR-MOB complexes, with a specific focus on the mechanistic basis for selective binding of MOB1 versus MOB2 to their respective kinase partners, a critical determinant in the functional output of Hippo and Hippo-like signaling pathways.
The NDR-MOB complex is characterized by a conserved architecture where the MOB coactivator binds the NTR region of the kinase, facilitating its activation and defining its functional role within cellular signaling networks.
The Conserved NTR-MOB Interface
The NDR kinase NTR domain adopts a V-shaped helical hairpin conformation upon binding to its MOB coactivator [1]. This structural motif is a common platform, observed in complexes from yeast to humans, including S. cerevisiae Cbk1–Mob2, Dbf2–Mob1, and human Lats1–Mob1 and Ndr–Mob1 [1]. The MOB protein itself possesses a highly conserved globular fold, a four alpha-helix bundle, which presents distinct surfaces for interaction with the kinase NTR and other regulatory proteins [15].
The primary role of MOB binding is to organize the NTR domain, enabling it to interact with the C-terminal hydrophobic motif (HM) of the kinase [1]. The HM is a critical regulatory element in AGC kinases, and in NDR/LATS kinases, its phosphorylation by upstream Hippo or Hippo-like kinases is essential for activation. The MOB-organized NTR mediates the association of the phosphorylated HM with an allosteric site on the kinase's N-terminal lobe, thereby stabilizing the active conformation of the kinase [1].
Table 1: Key Structural Features of Characterized NDR-MOB Complexes
| Complex | Organism | PDB Code(s) | Resolution (Å) | Key Structural Findings |
|---|---|---|---|---|
| Cbk1–Mob2 | S. cerevisiae | 4LQS, 4LQP, 4LQQ [2] | ~3.15 [1] | First full-length NDR/LATS-Mob structure; revealed novel coactivator-organized activation region and substrate docking mechanism [14]. |
| Cbk1NTR–Mob2 | S. cerevisiae | N/A | 2.8 [1] | Improved view of NTR-Mob2 interface; showed Mob2 organizes NTR to position the hydrophobic motif [1]. |
| Dbf2NTR–Mob1 | S. cerevisiae | N/A | 3.5 [1] | Provided a comparative structure to understand the basis of Mob binding specificity [1]. |
| Lats1NTR–hMob1 | H. sapiens | PDB 4RV9 [1] | N/A | Structural model of a human Hippo pathway core complex. |
Functional Consequences of MOB Binding
The formation of the NDR-MOB complex has several critical functional outcomes:
- Kinase Activation: MOB binding is essential for NDR/LATS kinase function. The association dramatically stimulates NDR1 and NDR2 catalytic activity [3].
- Allosteric Regulation: The complex creates a novel binding pocket that participates in the formation of the active state of the kinase after phosphorylation by upstream kinases [14].
- Substrate Docking: The structure of the Cbk1-Mob2 complex revealed a substrate docking mechanism previously unknown in AGC kinases, which provides robustness to the kinase's regulation of its in vivo substrates [14] [2].
Figure 1: MOB-Coordinated Kinase Activation Pathway. The MOB coactivator binds and organizes the NTR domain, which in turn mediates the positioning of the phosphorylated hydrophobic motif (HM) for allosteric kinase activation.
MOB1 vs. MOB2 Binding Specificity
A defining feature of NDR-MOB signaling is the high specificity of kinase-coactivator pairing. Generally, Lats kinases bind MOB1 proteins, while Ndr kinases bind MOB2 proteins, forming non-overlapping functional complexes in vivo [1] [16].
Structural Basis for Specificity
The specificity of MOB binding is not distributed across the entire interface but is restricted by discrete sites [1]. A short, variable motif within the MOB protein structure, which differs between MOB1 and MOB2, is a major determinant of this molecular recognition. Mutagenesis studies have shown that altering residues in the Cbk1 (Ndr) NTR allows association with the non-cognate MOB1 cofactor, confirming that specificity is governed by a limited set of interactions [1].
Functional Consequences of Specific Pairing
The specific MOB1-NDR and MOB2-NDR pairings lead to distinct functional outcomes:
- MOB1 Complexes: Typically activate kinases involved in mitotic exit and growth suppression, such as the Dbf2/20-Mob1 complex in the Mitotic Exit Network (MEN) and the LATS1/2-MOB1 complex in the metazoan Hippo pathway [14] [15].
- MOB2 Complexes: Typically activate kinases regulating cell polarity and morphogenesis, such as the Cbk1-Mob2 complex in the RAM network and the NDR1/2-MOB2 complex in animals [14].
In mammalian systems, the binding and functional outcomes can be more complex. hMOB2 binds to the N-terminal region of NDR1, but this binding differs significantly from hMOB1A. hMOB2 competes with hMOB1A for NDR binding and, unlike the activating hMOB1A, hMOB2 binding is associated with unphosphorylated NDR and functions as a negative regulator of NDR kinase activity [16]. RNAi depletion of hMOB2 results in increased NDR kinase activity, and its overexpression impairs NDR functions in apoptosis and centrosome duplication [16].
Table 2: Comparative Analysis of MOB1 vs. MOB2 Binding and Function
| Feature | MOB1 | MOB2 |
|---|---|---|
| Cognate Kinases | LATS subfamily (e.g., LATS1/2, Dbf2/20) [1] [14] | NDR subfamily (e.g., NDR1/2, Cbk1, SAX-1) [1] [17] |
| Primary Signaling Role | Hippo Pathway / MEN; growth control, mitotic exit [14] [15] | Hippo-like Pathway / RAM; morphogenesis, polarity, dendrite pruning [14] [17] |
| Effect on Kinase Activity | Activator [3] [15] | Context-dependent: Activator in yeast [14]; Competitor/Inhibitor of NDR in humans [16] |
| Key Specificity Determinant | Discrete short motif in MOB structure [1] | Discrete short motif in MOB structure [1] |
Figure 2: MOB-Kinase Binding Specificity and Competition. MOB1 and MOB2 typically form specific cognate complexes with LATS and NDR kinases, respectively. In human cells, MOB2 can compete with MOB1 for binding to NDR kinases and act as an inhibitor.
Experimental Protocols for Studying NDR-MOB Interactions
A multidisciplinary approach is required to dissect the structural and functional details of the NDR-MOB complex. The following protocols are based on methodologies successfully employed in the cited literature.
Protein Expression and Complex Purification
Objective: To produce high-quality, recombinant NDR and MOB proteins for structural and biochemical studies.
Detailed Protocol:
- Construct Design: Clone the DNA sequences encoding the NTR domain of the NDR kinase (e.g., Cbk1 residues 251-351) and the full-length MOB protein (e.g., Mob2) into compatible E. coli expression vectors, such as pGEX-4T1 or pMal-2c for generating GST- or MBP- fusion proteins, respectively [1] [16].
- Protein Stabilization (for unstable MOBs): For MOB proteins that are unstable in monomeric form (e.g., S. cerevisiae Mob2), engineer a stabilizing disulfide bond by introducing cysteine residues (e.g., Mob2 V148C Y153C) to mimic a zinc-binding motif found in metazoan orthologs [1].
- Co-expression and Purification:
- Co-express the His-tagged kinase NTR and the MOB protein in E. coli [1].
- Lyse cells and purify the complex using immobilized metal affinity chromatography (IMAC) via the His-tag.
- Further purify the complex by size-exclusion chromatography (SEC) to ensure homogeneity and remove aggregates. The complex typically elutes at a volume corresponding to a 1:1 heterodimer.
Crystallization and Structure Determination
Objective: To determine the high-resolution three-dimensional structure of the NDR-MOB complex.
Detailed Protocol:
- Crystallization: Use the vapor-diffusion method. Set up trials with the purified complex at concentrations of 10-20 mg/mL. Crystals of the Cbk1NTR–Mob2 complex were obtained in conditions such as 0.1M sodium citrate tribasic dihydrate pH 5.5, 15% v/v 1,4-Dioxane, 15% w/v PEG-20,000 [1].
- Data Collection and Processing: Flash-cool crystals in liquid nitrogen using a cryoprotectant. Collect X-ray diffraction data at a synchrotron beamline. Process the data (indexing, integration, and scaling) using software like XDS or HKL-2000 [1].
- Structure Solution: Solve the phase problem by molecular replacement (MR) using a known MOB structure (e.g., PDB 4JIZ) and an NTR model as search models. Iterative model building and refinement are performed using Coot and Phenix/Refmac [1] [14].
Table 3: Crystallographic Data Collection and Refinement Statistics (Example from [1])
| Parameter | Cbk1NTR–Mob2 | Dbf2NTR–Mob1 |
|---|---|---|
| Data Collection | ||
| Space Group | P41212 | P6122 |
| Resolution (Å) | 39.9 - 2.8 | 46.9 - 3.5 |
| Refinement | ||
| Rwork / Rfree | 0.2490 / 0.2838 | 0.2292 / 0.2631 |
| No. of Atoms | 2196 | 2024 |
| B-factors (Ų) | 92.7 | 88.3 |
| Ramachandran Plot | ||
| - Favored (%) | 93 | 93.5 |
| - Outliers (%) | 3 | 1.6 |
Functional Analysis of Binding Specificity and Kinase Activity
Objective: To validate the structural findings and quantify the functional impact of NDR-MOB interactions.
Detailed Protocol:
- Site-Directed Mutagenesis: Generate point mutations in the specificity-determining motifs of the NTR (e.g., Cbk1 NTR) or the MOB protein to test their effect on binding and specificity [1].
- Binding Affinity Measurements:
- Co-immunoprecipitation (Co-IP): Express wild-type and mutant proteins in mammalian cells (e.g., COS-7, HEK 293). Immunoprecipitate the kinase and probe for co-precipitating MOB protein by western blotting [16].
- Quantitative Binding Assays: Use techniques like Surface Plasmon Resonance (SPR) or Isothermal Titration Calorimetry (ITC) to determine the binding affinity (KD) between purified wild-type and mutant NTR and MOB proteins.
- Kinase Activity Assays: Measure the activity of the full-length NDR kinase in the presence of its cognate or non-cognate MOB partner. Use in vitro kinase assays with a suitable substrate (e.g., myelin basic protein) and [γ-32P]ATP. Quantify phosphate incorporation to assess the stimulatory or inhibitory effect of MOB binding [16] [3].
The Scientist's Toolkit: Key Research Reagents
Table 4: Essential Reagents for Investigating NDR-MOB Complexes
| Reagent / Tool | Function / Application | Example & Notes |
|---|---|---|
| Stabilized MOB Variants | Enables structural studies of unstable MOB proteins. | S. cerevisiae Mob2 V148C Y153C (zinc-binding variant) [1]. |
| Kinase Activation Reporters | Measures NDR/LATS kinase activity in vitro and in cells. | Phospho-specific antibodies against NDR HM (e.g., pT444/T442); generic kinase substrates (e.g., Myelin Basic Protein) [16]. |
| Specificity Mutants | Probes the structural basis of selective MOB binding. | Mutations in the short specificity motif of the NTR (e.g., Cbk1 NTR mutants) or MOB protein [1]. |
| Tet-Inducible Expression Vectors | Allows controlled overexpression of MOB proteins in mammalian cells for functional studies. | pT-Rex-DEST30 vectors for inducible expression of hMOB2 [16]. |
| RNAi Knockdown Systems | Assesses the functional consequences of depleting specific MOBs. | pTER vectors expressing shRNA against hMOB2 to study its role as a negative regulator [16]. |
| Molecular Dynamics (MD) Software | Models the dynamics and conformational changes of the NDR-MOB complex. | Used to simulate HM engagement and allosteric activation [14]. |
Molecular Determinants of MOB1 Specificity for LATS Kinases
The Mps One Binder (MOB) family of adapter proteins represents crucial regulatory components in key signaling pathways, including the Hippo tumor suppressor network. While MOB proteins share structural homology, they exhibit distinct binding specificities for members of the NDR/LATS kinase family. This technical analysis examines the molecular determinants governing MOB1's specific recognition of LATS kinases over the closely related NDR kinases. Through structural, biochemical, and genetic perspectives, we delineate how specific residue interactions, phosphorylation events, and allosteric mechanisms confer binding specificity. Understanding these precise molecular interactions provides critical insights for targeted therapeutic interventions in cancers where Hippo pathway dysregulation plays a fundamental role.
MOB proteins constitute an evolutionarily conserved family of signal transducers that function as essential coactivators of AGC group kinases, particularly the NDR/LATS subfamily [13] [18]. In humans, six MOB proteins (MOB1A/B, MOB2, MOB3A/B/C, MOB4) have been identified, with MOB1A and MOB1B sharing 95% sequence identity and functioning redundantly in Hippo pathway signaling [18]. MOB1 has emerged as a pivotal integrator within the core kinase cassette of the Hippo pathway, binding both upstream MST1/2 kinases and downstream LATS1/2 kinases to facilitate signal transduction [19] [20].
The specificity of MOB proteins for their kinase partners represents a critical regulatory node in cellular signaling. While MOB1 interacts with both LATS and NDR kinases, MOB2 demonstrates exclusive binding to NDR kinases, creating distinct functional branches [4]. This specificity is not merely a binary interaction but governs fundamental biological processes including cell proliferation, apoptosis, organ size control, and tumor suppression [18] [21]. Disruption of specific MOB1-LATS interactions has been directly linked to tumorigenesis, highlighting the physiological importance of precise molecular recognition [18].
Table 1: MOB Family Binding Specificities for NDR/LATS Kinases
| MOB Protein | NDR1/2 Binding | LATS1/2 Binding | Primary Functions |
|---|---|---|---|
| MOB1A/B | Yes | Yes | Hippo pathway core signaling, tumor suppression |
| MOB2 | Yes | No | Cell cycle regulation, DNA damage response |
| MOB3A/B/C | No | No | Binds MST1, apoptotic regulation? |
| MOB4 | Unknown | Unknown | Poorly characterized |
Structural Basis of MOB-Kinase Interactions
Structural analyses of MOB1 in complex with its kinase partners have revealed conserved yet distinct binding modes. The crystal structure of MOB1 adopts a globular shape consisting of nine α-helices (α1–α9) and two β-strands, forming a characteristic phosphopeptide-binding pocket and kinase interaction surface [18] [22]. Comparative analysis of MOB1 bound to LATS1 versus NDR2 reveals that both kinases bind MOB1 through a V-shaped structure composed of two antiparallel α-helices that engage with negatively charged electrostatic surfaces on MOB1 [18].
The determination of the MOB1/NDR2 complex structure at 2.1 Å resolution enabled direct comparison with existing MOB1/LATS1 structures, revealing both shared core interaction principles and critical differences that underlie specificity [18]. In both complexes, the N-terminal regulatory domains (NTR) of the kinases interact with overlapping but distinct surfaces on MOB1 through a combination of hydrogen bonds, van der Waals interactions, and electrostatic complementarity [18] [22].
The Critical Asp63 Determinant for LATS Specificity
Comparative structural analysis between MOB1/NDR2 and MOB1/LATS1 complexes revealed a key specificity determinant at position Asp63 of MOB1 [18] [22]. This residue forms a specific bond with His646 of LATS1, supported by a cluster of surrounding residues involving Phe642, Met643, Gln645, Val647, and Val650 [22]. In contrast, Phe31 of NDR2 does not interact with Asp63 of MOB1, explaining the lack of this specific stabilizing interaction in the MOB1/NDR complex [18].
Mutagenesis studies confirm the functional importance of this interaction, where MOB1-D63A mutation selectively disrupts LATS1 binding while preserving NDR2 interaction [18]. This single residue thereby serves as a molecular switch governing kinase partner specificity, with profound functional consequences for Hippo pathway signaling and tumor suppressor activity.
Table 2: Key Residues in MOB1-Kinase Interfaces
| Complex | MOB1 Residues | Kinase Residues | Interaction Type | Functional Significance |
|---|---|---|---|---|
| MOB1/LATS1 | Asp63, Glu51, Glu55, Trp56, Val59 | His646, Phe642, Met643, Gln645, Val647 | Hydrogen bonding, van der Waals | LATS-specific binding, essential for tumor suppression |
| MOB1/NDR2 | Leu36, Gly39, Leu41, Ala44, Gln67 | Lys25, Leu28, Tyr32, Leu35, Ile36 | Hydrophobic interactions, van der Waals | Conserved NDR/LATS binding |
| MOB1/MST1/2 | K153, R154, R157 | Phospho-Thr/Ser motifs (e.g., pT353, pT367) | Phosphopeptide recognition | Upstream regulation, pathway activation |
Biochemical and Functional Validation
Quantitative Binding Analyses
Systematic biochemical analyses have quantified the binding affinities and specificities of MOB1 for its kinase partners. Fluorescence polarization binding experiments with purified components demonstrate that MOB1 binds LATS1 with approximately 5-10 fold higher affinity compared to NDR2, with dissociation constants in the low micromolar range [19] [20]. This enhanced affinity for LATS1 is largely abolished in MOB1-D63A mutants, confirming the structural observations at a quantitative level [18].
Phosphorylation events further modulate these interactions. MOB1 phosphorylation by MST1/2 increases its ability to activate LATS1/2 approximately 3-fold, creating a positive feedback loop that enhances pathway signaling [19] [20]. This phosphorylation-dependent enhancement is less pronounced for NDR kinase activation, suggesting additional layers of specificity regulation.
Functional Consequences in Cellular Contexts
Genetic studies utilizing MOB1 variants with selective loss-of-interaction have demonstrated the functional importance of specific MOB1-LATS binding. In human cancer cells, disruption of MOB1-LATS interaction (but not MOB1-NDR interaction) abrogates the tumor-suppressive properties of MOB1, including growth inhibition and YAP/TAZ phosphorylation [18].
Drosophila genetics corroborate these findings, demonstrating that the MOB1/Warts (LATS homolog) interaction is essential for development and tissue growth control, while stable MOB1/Hippo (MST homolog) binding is dispensable [18] [22]. This in vivo evidence establishes the MOB1-LATS interaction as the critical node for Hippo pathway tumor suppressor function.
Experimental Approaches for Characterizing MOB1 Specificity
Structural Biology Methodologies
Protein Expression and Purification: Recombinant MOB1 and kinase N-terminal domains (e.g., NDR2 25-88, LATS1 homologous region) are expressed in E. coli BL21(DE3) as N-terminal dual 6xhistidine and glutathione S-transferase (GST) fusion proteins using modified pETM-30 vectors [19]. Proteins are purified using glutathione-Sepharose resin, followed by TEV protease cleavage to remove affinity tags, and final purification by size exclusion chromatography.
Crystallization and Structure Determination: Complexes of MOB1 with kinase peptides are formed by mixing at 1:1.5 mole ratio. Crystals are obtained by vapor diffusion using hanging drops with 1:1 mixtures of protein (7 mg/ml concentration) with precipitant solution (0.1 M MES pH 6.0, 0.2 M LiCl, 20% PEG 6000) [19]. X-ray diffraction data are collected at synchrotron sources (e.g., Northeastern Collaborative Access Team beamlines), and structures solved by molecular replacement.
Biochemical Interaction Assays
Fluorescence Polarization Binding Assays: FITC-labeled phosphopeptides corresponding to MOB1-binding motifs in MST1/2 (e.g., T353, T367 peptides) are used for quantitative binding measurements [19]. Serial dilutions of MOB1 proteins are incubated with fixed concentrations of labeled peptides, and polarization values measured to determine dissociation constants.
GST Pull-Down and Far-Western Analyses: For protein-protein interaction studies, GST-tagged MOB1 variants are immobilized on glutathione-Sepharose resin and incubated with potential binding partners [19] [18]. After washing, bound proteins are eluted and detected by immunoblotting. For Far-Western analysis, proteins are separated by SDS-PAGE, transferred to membranes, and probed with purified MOB1 proteins followed by anti-MOB1 antibodies.
Research Reagent Solutions
Table 3: Essential Research Reagents for MOB1-LATS Specificity Studies
| Reagent Category | Specific Examples | Function/Application | Key Features |
|---|---|---|---|
| Expression Vectors | pETM-30 (modified) | Recombinant protein expression | Dual 6xHis-GST tags, TEV cleavage site |
| MOB1 Variants | MOB1A wild-type, MOB1-D63A, MOB1 phosphomimetics | Structure-function studies | Selective binding defects, modified regulation |
| Kinase Constructs | NDR2 NTR (25-88), LATS1 NTR, full-length kinases | Interaction partners | Defined binding domains, functional assays |
| Phosphopeptides | MST1 pT353, MST1 pT367, optimized Nud1-like | Binding specificity studies | FITC-labeled for quantitative assays |
| Crystallization Reagents | MES pH 6.0, LiCl, PEG 6000, ethylene glycol | Structural biology | Crystal optimization, cryoprotection |
| Detection Tools | Anti-phospho-NDR/LATS antibodies, anti-MOB1 antibodies | Functional validation | Pathway activation status, complex formation |
Discussion: Implications for Therapeutic Targeting
The precise molecular determinants governing MOB1 specificity for LATS kinases represent attractive targets for therapeutic intervention in cancers characterized by Hippo pathway dysregulation. The identification of Asp63 as a key residue for LATS-specific binding suggests that small molecules mimicking or disrupting this interaction could selectively modulate Hippo signaling branch outcomes.
Furthermore, the phosphoregulation of MOB1 interactions offers additional opportunities for pharmacological manipulation. As MOB1 phosphorylation enhances LATS activation, strategies to promote this specific phosphorylation event could potentiate tumor suppressor activity in contexts where pathway activity is diminished [19] [20]. Conversely, in conditions of excessive pathway activation, targeted disruption of MOB1-LATS complex formation might provide therapeutic benefit.
The differential binding properties of MOB1 versus MOB2 also suggest that tissue-specific expression patterns of these adapters could create natural specificities that might be exploited for targeted therapies with reduced off-target effects [4]. As our structural understanding of these complexes advances, structure-based drug design approaches become increasingly feasible for developing next-generation cancer therapeutics targeting the Hippo pathway.
The molecular determinants of MOB1 specificity for LATS kinases encompass a sophisticated integration of structural complementarity, specific residue interactions, and regulatory phosphorylation events. The Asp63-His646 interaction emerges as a critical specificity switch that distinguishes LATS from NDR binding, with profound functional consequences for tumor suppression and tissue growth control. Continued structural and biochemical dissection of these interfaces, coupled with functional validation in physiological contexts, will further refine our understanding of this crucial signaling node and its therapeutic potential.
Key Structural Motifs Governing MOB2 Binding to NDR1/2
The Mps one binder (MOB) proteins are highly conserved eukaryotic signal transducers that function as crucial regulatory cofactors for the NDR/LATS family of serine/threonine kinases [4]. In humans, the MOB family consists of six members (MOB1A, MOB1B, MOB2, MOB3A, MOB3B, and MOB3C) that exhibit distinct binding specificities toward the NDR/LATS kinases [16]. MOB2 displays remarkable specificity for the NDR1 and NDR2 kinases (also known as STK38 and STK38L, respectively), in contrast to MOB1, which can bind to both NDR1/2 and LATS1/2 kinases [16] [23]. This review focuses on the key structural motifs governing MOB2 binding to NDR1/2, framed within the broader context of MOB1 versus MOB2 binding specificity. Understanding these molecular determinants is essential for elucidating how specific MOB-NDR complexes control diverse cellular processes, including cell cycle progression, the DNA damage response, and cell motility [4] [23]. The competitive relationship between MOB1 and MOB2 for NDR binding represents a critical regulatory mechanism for Hippo pathway signaling, with significant implications for both fundamental biology and therapeutic development [16].
Structural Organization of NDR Kinases and MOB Cofactors
Domain Architecture of NDR Kinases
NDR1 and NDR2 are serine-threonine kinases belonging to the AGC kinase family that share approximately 87% sequence identity [3] [8]. Despite their high similarity, they exhibit distinct subcellular localizations—NDR1 is primarily nuclear, while NDR2 displays a punctate cytoplasmic distribution—suggesting non-redundant cellular functions [3]. Beyond the conserved kinase domain characteristic of AGC kinases, NDR kinases possess two critical regulatory regions: an N-terminal regulatory (NTR) region and a C-terminal hydrophobic motif (HM) [1] [24]. The NTR forms a structural platform specifically dedicated to MOB protein binding, while the HM contains a threonine residue (T444 in NDR1) whose phosphorylation by upstream kinases is essential for NDR activation [1] [24].
Structural Features of MOB Proteins
MOB proteins adopt a conserved globular fold known as the Mob1/Phocein domain, which consists of a core β-sandwich structure formed by two β-sheets [1] [2]. Despite this common structural framework, MOB1 and MOB2 possess distinct surface properties that dictate their binding specificities for different NDR/LATS kinases. Structural analyses reveal that while MOB1 can bind both NDR and LATS kinases, MOB2 exhibits specific binding only to NDR1/2 kinases [16] [23]. This specificity is mediated by discrete molecular recognition motifs rather than broadly distributed interface properties [1].
Table 1: Key Structural Features of MOB Proteins and NDR Kinases
| Component | Key Structural Features | Functional Role |
|---|---|---|
| NDR Kinase NTR | V-shaped helical hairpin [1] | MOB protein binding platform |
| NDR Kinase HM | C-terminal extension with phosphorylatable threonine [24] | Kinase activation through allosteric regulation |
| MOB Protein Core | Conserved globular β-sandwich fold [2] | Scaffold for kinase interaction |
| MOB Specificity Determinants | Discrete surface motifs [1] | Dictate binding preference for NDR vs. LATS kinases |
Molecular Basis of MOB2-NDR1/2 Interactions
The NTR-MOB Interface: A Structural Platform for Kinase Regulation
The primary interaction between MOB2 and NDR1/2 occurs through the N-terminal regulatory (NTR) region of the kinases, which forms a V-shaped helical hairpin that docks against a complementary surface on MOB2 [1]. Structural studies of the Saccharomyces cerevisiae Cbk1 (NDR homolog)-Mob2 complex, which serves as a model for understanding human NDR-MOB interactions, reveal that the NTR-Mob interface provides a distinctive kinase regulation mechanism [1] [2]. In this complex, the MOB2 cofactor organizes the NDR NTR to interact with the AGC kinase C-terminal hydrophobic motif (HM), facilitating its positioning for allosteric regulation [1]. This MOB-organized NTR appears to mediate association of the HM with an allosteric site on the N-terminal kinase lobe, creating an integrated regulatory system unique to NDR/LATS kinases [1].
High-resolution crystal structures of the Cbk1NTR–Mob2 complex show that the NTR forms a bihelical conformation similar to those observed in LatsNTR– and NdrNTR–Mob1 structures, indicating evolutionary conservation of this structural platform [1]. The improved resolution of these recent structures (2.8 Å) compared to earlier models has highlighted the critical role of MOB binding in positioning the hydrophobic motif of the kinase, revealing how MOB-driven orientation promotes optimal positioning of the kinase's otherwise flexible αC helix, a component critical for kinase activation [1].
Key Residues and Motifs Governing Binding Specificity
The specificity of MOB2 for NDR1/2 kinases, as opposed to the broader binding capacity of MOB1, is mediated by discrete molecular recognition sites. Research indicates that alteration of specific residues in the Cbk1 NTR allows association with the noncognate Mob1 cofactor, demonstrating that cofactor specificity is restricted by discrete sites rather than being broadly distributed across the interaction surface [1]. These specificity determinants include a short motif in the Mob structure that differs between Mob1 and Mob2, strongly contributing to molecular recognition between the kinase and cofactor [1].
Biochemical studies demonstrate that MOB2 competes with MOB1 for binding to the same N-terminal region of NDR1, but with fundamentally different functional outcomes [16]. While MOB1 binding activates NDR kinases by stimulating autophosphorylation on the activation segment, MOB2 associates predominantly with unphosphorylated NDR and is associated with diminished NDR kinase activity [16]. This competitive relationship creates a regulatory switch where the relative abundance and activation state of MOB1 versus MOB2 can determine NDR kinase activity levels.
Table 2: Functional Consequences of MOB1 vs. MOB2 Binding to NDR1/2
| Parameter | MOB1-NDR Complex | MOB2-NDR Complex |
|---|---|---|
| NDR Kinase Activity | Increased [16] | Diminished [16] |
| NDR Phosphorylation State | Preferentially binds phosphorylated NDR [16] | Preferentially binds unphosphorylated NDR [16] |
| Competitive Binding | Displaced by MOB2 overexpression [16] | Competes with MOB1 for NDR binding [16] |
| Cellular Function | Promotes NDR-mediated signaling [4] | May buffer or modulate NDR signaling [4] |
MOB1 vs. MOB2: A Comparative Analysis of Binding Specificity
Structural Determinants of Differential Binding
While both MOB1 and MOB2 bind to the NTR region of NDR kinases, structural and biochemical evidence indicates their binding modes differ significantly [16]. The human MOB1-NDR2 complex structure (PDB: 5XQZ) reveals specific interactions mediated by key MOB1 residues that enable its differential binding to Hippo core kinases [25]. Comparative analyses suggest that MOB2 lacks certain structural features present in MOB1 that are necessary for stable interaction with LATS kinases, explaining its restriction to NDR kinases [16] [23].
The specificity of MOB-NDR/LATS interactions is evolutionarily conserved. In budding yeast, the NDR-subfamily kinase Cbk1 associates specifically with Mob2, while the LATS-related kinases Dbf2 and Dbf20 bind exclusively to Mob1 [1]. This specific pairing is maintained despite the simultaneous presence of all proteins in the cytosol, indicating a robust mechanism enforcing kinase-coactivator association specificity [1]. Studies of homologous Schizosaccharomyces pombe Sid2(Lats)-Mob1 and Orb6(Ndr)-Mob2 complexes further demonstrate highly specific NDR/LATS-Mob interactions, preserved across vast evolutionary distances [1].
Functional Consequences of Distinct MOB Binding
The differential binding of MOB1 and MOB2 to NDR kinases has significant functional implications. MOB1 binding activates NDR1/2 kinases by stimulating autophosphorylation on the activation segment and facilitates phosphorylation by upstream kinases such as MST1 [16] [20]. In contrast, MOB2 binding is associated with diminished NDR activity, and RNA interference-mediated depletion of MOB2 results in increased NDR kinase activity, indicating that MOB2 functions as a negative regulator of human NDR kinases [16].
This competitive regulation extends to biological functions. Overexpression of MOB2 interferes with NDR roles in death receptor signaling and centrosome duplication, consistent with its function as a competitive inhibitor of MOB1-NDR complex formation [16]. Furthermore, research suggests that MOB2 regulates the alternative interaction of MOB1 with NDR1/2 and LATS1, leading to increased phosphorylation of LATS1 and MOB1 and consequent inactivation of YAP, ultimately inhibiting hepatocellular carcinoma cell motility [23].
Experimental Approaches for Studying MOB2-NDR Interactions
Structural Biology Methodologies
Determining the molecular details of MOB2-NDR interactions has required sophisticated structural biology approaches. X-ray crystallography has been instrumental, with structures of complexes such as Cbk1NTR–Mob2 determined to 2.8 Å resolution [1]. These studies often require protein engineering to enhance stability; for instance, introducing a zinc-binding motif (Mob2 V148C Y153C) stabilizes Mob2 for suitable Escherichia coli expression [1].
Crystallographic data collection typically involves several steps as shown in the workflow below:
Diagram 1: Structural Biology Workflow for MOB-NDR Complex Analysis
Biochemical and Cellular Assays
Beyond structural approaches, researchers employ diverse biochemical and cellular assays to characterize MOB2-NDR interactions. Coimmunoprecipitation experiments demonstrate physical association between MOB2 and NDR kinases in cellular contexts [16] [3]. Kinase activity assays show that MOB2 association dramatically stimulates NDR1 and NDR2 catalytic activity, in contrast to its competitive regulatory role with MOB1 [3].
Functional studies often utilize RNA interference-mediated depletion of MOB2, which results in increased NDR kinase activity, supporting MOB2's role as a negative regulator [16]. Overexpression approaches further reveal that MOB2 impairs NDR1/2 activation in a binding-dependent manner and affects NDR functions in centrosome duplication and apoptotic signaling [16]. More recently, CRISPR/Cas9-mediated knockout of MOB2 has been employed to investigate its role in hepatocellular carcinoma cell migration and invasion, demonstrating that MOB2 knockout promotes these processes while decreasing phosphorylation of YAP [23].
Table 3: Key Experimental Reagents for Studying MOB2-NDR Interactions
| Reagent/Solution | Function/Application | Experimental Context |
|---|---|---|
| pcDNA3 vectors with epitope tags | Mammalian expression of MOB/NDR proteins [16] | Immunoprecipitation, localization studies |
| pGEX-4T1/pMal-2c vectors | Bacterial expression for protein purification [16] | Structural studies, in vitro binding assays |
| pTER-shMOB2 vectors | RNAi-mediated MOB2 knockdown [16] | Functional analysis of MOB2 depletion |
| LentiCRISPRv2 vector | CRISPR/Cas9-mediated MOB2 knockout [23] | Generation of stable knockout cell lines |
| Tet-regulated expression vectors | Inducible MOB2 expression [16] | Controlled overexpression studies |
Implications for Cellular Signaling and Therapeutic Development
Roles in Cell Cycle Regulation and DNA Damage Response
The MOB2-NDR interaction plays significant roles in cell cycle progression and DNA damage response. MOB2 depletion triggers a p53/p21-dependent G1/S cell cycle arrest in untransformed human cells, suggesting its importance in cell cycle checkpoint control [4]. Furthermore, MOB2 is required to prevent the accumulation of endogenous DNA damage and prevent undesired activation of cell cycle checkpoints [4]. Intriguingly, these functions may operate independently of NDR1/2 kinase signaling, as NDR1 or NDR2 knockdown does not recapitulate the cell cycle arrest phenotype observed with MOB2 depletion [4].
MOB2 also functions as a novel DNA damage response (DDR) factor that plays roles in DDR signaling, cell survival, and cell cycle checkpoints upon exposure to DNA damage [4]. It is required to promote cell survival and G1/S cell cycle arrest upon exposure to DNA damaging agents such as ionizing radiation or doxorubicin [4]. MOB2 supports ionizing radiation-induced DDR signaling through the DDR kinase ATM and facilitates recruitment of the MRE11-RAD50-NBS1 (MRN) DNA damage sensor complex and activated ATM to DNA damaged chromatin [4].
Relevance to Cancer and Therapeutic Opportunities
The MOB2-NDR interaction has significant implications for cancer biology and therapeutic development. In hepatocellular carcinoma, MOB2 inhibits cell motility by regulating the alternative interaction of MOB1 with NDR1/2 and LATS1, resulting in increased phosphorylation of LATS1 and MOB1 and consequent inactivation of YAP [23]. This positions MOB2 as a potential tumor suppressor in specific contexts.
The diagram below illustrates how MOB2 integrates into Hippo signaling and impacts cancer-relevant processes:
Diagram 2: MOB2 Integration in Hippo Signaling and Cancer
Given NDR2's role as an oncogene in most cancers—particularly lung cancer, where it regulates processes including proliferation, apoptosis, migration, invasion, and vesicular trafficking—the MOB2-NDR interface represents a potential therapeutic target [8]. The specific structural motifs governing MOB2-NDR binding could be exploited for developing targeted protein-protein interaction inhibitors that modulate this signaling axis in cancer therapy.
The structural motifs governing MOB2 binding to NDR1/2 represent a sophisticated molecular recognition system that controls specific signaling outputs within the broader Hippo pathway framework. The NTR region of NDR kinases serves as a structural platform that binds MOB cofactors, with discrete molecular determinants dictating the specificity of MOB2 for NDR kinases versus the broader binding capacity of MOB1. This specific interaction, characterized by competitive binding with MOB1 and association with unphosphorylated NDR, positions MOB2 as a negative regulator of NDR kinase activity. The functional consequences of MOB2-NDR interactions extend to diverse cellular processes, including cell cycle regulation, DNA damage response, and control of cell motility in cancer contexts. Future research elucidating the full complement of regulatory inputs and outputs of the MOB2-NDR axis will undoubtedly yield valuable insights for therapeutic intervention in cancer and other diseases.
The hydrophobic motif (HM) represents a critical regulatory element in the C-terminal tail of AGC family protein kinases, serving as a central interface for controlling kinase activity, substrate recognition, and integration into cellular signaling networks. This conserved motif functions as a molecular switch that governs allosteric activation through phosphorylation-dependent mechanisms and protein-protein interactions. Within the NDR/LATS kinase subfamily, the HM serves as a pivotal regulatory hub that integrates signals from upstream kinases, coactivator proteins, and scaffolding elements to control diverse cellular processes including cell division, morphogenesis, and proliferation [26] [1].
The significance of HM regulation extends beyond fundamental biology to therapeutic applications, as evidenced by the critical role of HM phosphorylation in AKT activation, a prominent oncogenic signaling pathway [27]. This technical review examines the structural and mechanistic principles of HM function, with particular emphasis on the molecular determinants governing MOB coactivator binding specificity in NDR kinase regulation. Through integrated analysis of quantitative biochemical data, structural insights, and experimental methodologies, we provide a comprehensive framework for understanding HM-mediated kinase control and its implications for targeted therapeutic development.
Structural and Mechanistic Principles of Hydrophobic Motif Function
Consensus Features and Structural Location
The hydrophobic motif is characterized by a conserved sequence profile typically containing a phosphorylation site (most commonly a threonine residue) embedded within a hydrophobic context. In NDR kinases, this motif is located C-terminal to the kinase catalytic domain and adopts specific structural configurations that are stabilized by phosphorylation-dependent interactions. Structural analyses reveal that the phosphorylated HM engages in intramolecular interactions with the N-lobe of the kinase domain, particularly stabilizing the αC-helix in an active conformation [1] [24].
This HM-mediated stabilization is essential for proper alignment of catalytic residues and formation of the regulatory (R) and catalytic (C) spines that traverse the kinase core. The NDR/LATS kinase family exhibits a unique structural adaptation where the Mob coactivator organizes the N-terminal regulatory (NTR) region to mediate interaction between the phosphorylated HM and an allosteric site on the N-terminal kinase lobe [1]. This configuration creates a distinctive kinase-coactivator system that integrates HM phosphorylation with coactivator binding for full kinase activation.
Phosphorylation-Dependent Activation Mechanisms
HM phosphorylation triggers a conformational transition from an autoinhibited state to an active kinase configuration. In NDR1, structural studies have identified an atypically long activation segment that autoinhibits the kinase domain by blocking substrate binding and stabilizing the αC-helix in a non-productive position [24]. Phosphorylation of the HM residue (Thr444 in NDR1, Thr442 in NDR2) relieves this autoinhibition and enables the conformational rearrangements necessary for catalytic competence.
The activation mechanism involves a multi-step process wherein HM phosphorylation enhances the efficiency of activation loop phosphorylation and stabilizes the active conformation against phosphatases. This cooperative activation creates a switch-like response to upstream signals and ensures precise temporal control of kinase activity. In NDR kinases, this process is further refined through interaction with MOB coactivators, which potentiate kinase activity through mechanisms distinct from HM phosphorylation [24].
Table 1: Hydrophobic Motif Characteristics in Selected AGC Kinases
| Kinase | HM Sequence | Phosphorylation Site | Upstream Regulator | Functional Consequences |
|---|---|---|---|---|
| NDR1 | FXXT444 | Thr444 | MST3 | 10-fold activation; enhanced MOB1A binding [26] |
| NDR2 | FXXT442 | Thr442 | MST3 | Conformational activation; cytoplasmic localization [28] |
| AKT1 | FPQFS473 | Ser473 | mTORC2 | Stabilizes Thr308 phosphorylation; maximal activity [27] |
| Cbk1 | FXXT743 | Thr743 | Unknown | Promotes HM positioning via Mob2-organized NTR [1] |
MOB Coactivator Binding Specificity in NDR Kinase Regulation
Structural Basis of MOB-NDR Interactions
The NDR/LATS kinases possess a distinctive N-terminal regulatory (NTR) region that forms a highly specific interface with Mob coactivator proteins. Structural analyses of Saccharomyces cerevisiae Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes reveal that the NTR forms a V-shaped helical hairpin that docks against the conserved surface of Mob coactivators [1]. This interface serves as a structural platform that mediates kinase-cofactor binding and enables allosteric regulation of the HM.
The specificity determinants that govern selective association of Ndr kinases with Mob2 (versus Lats kinases with Mob1) reside in discrete structural elements rather than being broadly distributed across the interaction surface. In Cbk1, alteration of specific residues in the NTR allows association with the non-cognate Mob1 cofactor, indicating that specificity is controlled by molecular complementarity at restricted sites [1]. This precise molecular recognition ensures proper pathway specificity despite the high conservation of both the Mob cofactors and kinase NTR regions across the NDR/LATS family.
Functional Integration of HM Phosphorylation and MOB Binding
The activation of NDR kinases requires the functional integration of HM phosphorylation with MOB coactivator binding. Research demonstrates that MST3-mediated phosphorylation of Thr442 in NDR2 results in approximately 10-fold stimulation of kinase activity, while subsequent MOB1A binding further increases activity, leading to a fully active kinase [26]. This cooperative activation creates a multi-step mechanism that enables signal integration and precise control of NDR kinase function.
Structural evidence indicates that MOB binding organizes the NTR to interact with the phosphorylated HM, thereby facilitating optimal positioning of the otherwise flexible αC-helix critical for kinase activation [1]. This mechanism explains the essential role of both HM phosphorylation and MOB coactivator binding for NDR kinase function, and illustrates how these two regulatory inputs are structurally coupled through the NTR-Mob interface.
Table 2: MOB Cofactor Specificity and Functional Roles in NDR/LATS Kinases
| Kinase | MOB Cofactor | Binding Specificity Determinants | Functional Consequences | Biological Context |
|---|---|---|---|---|
| Cbk1 (NDR) | Mob2 | Discrete sites in NTR; short motif in Mob structure | Organizes NTR for HM positioning; substrate docking [1] | RAM network; cell separation, morphogenesis [2] |
| Dbf2 (LATS) | Mob1 | Molecular complementarity at restricted interface | Kinase coactivation; spindle pole recruitment [1] | Mitotic Exit Network (MEN); cytokinesis [2] |
| SAX-1/NDR | MOB-2 | Conserved interface with helical hairpin NTR | Dendrite pruning; membrane dynamics regulation [17] | Neuronal remodeling; C. elegans [17] |
Experimental Analysis of Hydrophobic Motif Regulation
Methodologies for Assessing HM Phosphorylation
The investigation of HM phosphorylation employs multiple complementary approaches to establish regulatory mechanisms:
In Vitro Kinase Assays: Recombinant MST3 kinase selectively phosphorylates Thr442 of NDR2 in vitro, resulting in significant kinase activation. These assays typically use purified components under controlled conditions to establish direct phosphorylation relationships [26].
Phosphospecific Antibodies: Antibodies specifically recognizing phosphorylated HM residues (e.g., anti-P-Thr-442 for NDR2) enable quantitative assessment of HM phosphorylation status in both in vitro and cellular contexts [26] [28].
Kinase-Dead Mutants: MST3KR, a kinase-dead mutant of MST3, potently inhibits Thr442 phosphorylation following okadaic acid stimulation in vivo, establishing the functional requirement for MST3 catalytic activity [26].
Knockdown Approaches: Short hairpin RNA constructs targeting MST3 effectively abolish Thr442 hydrophobic motif phosphorylation in HEK293F cells, demonstrating necessity in a cellular context [26].
Structural Biology Techniques
High-resolution structural biology provides essential insights into HM regulatory mechanisms:
X-ray Crystallography: Crystal structures of NDR1 kinase domain (2.2 Å resolution) reveal the autoinhibitory function of an atypically long activation segment that blocks substrate binding and stabilizes the αC-helix in a non-productive position [24].
Complex Structure Determination: Structures of Cbk1NTR–Mob2 (2.8 Å resolution) and Dbf2NTR–Mob1 complexes elucidate the molecular basis of coactivator binding specificity and HM positioning [1].
Mutational Analysis: Structure-guided mutations within the activation segment of NDR1 dramatically enhance in vitro kinase activity, confirming autoinhibitory function [24].
Diagram 1: HM Phosphorylation Analysis Workflow. This experimental workflow outlines key methodological steps for assessing hydrophobic motif phosphorylation status, incorporating specific reagents and conditions from cited studies [26].
Research Reagent Solutions for HM Regulation Studies
Table 3: Essential Research Reagents for Investigating HM Regulation
| Reagent Category | Specific Examples | Experimental Function | Application Context |
|---|---|---|---|
| Phosphospecific Antibodies | Anti-P-Thr-442 (NDR2); Anti-P-Ser-282 (NDR2) | Detect HM and activation loop phosphorylation | Western blotting; monitoring cellular phosphorylation status [26] |
| Kinase Constructs | HA-NDR2; myc-MST3; HA-MST3KR (kinase-dead) | Functional analysis of kinase activity and regulation | Transfection studies; in vitro kinase assays [26] [29] |
| Mob Coactivators | myc-C1-MOB1A; pGEX-2T-MOB1A | Investigate coactivator-kinase functional interactions | Co-immunoprecipitation; kinase activation assays [26] |
| Expression Vectors | pCMV5-HA-NDR2; pTER-shMST3 (shRNA) | Modulate protein expression in cellular systems | Knockdown studies; heterologous protein expression [26] |
| Chemical Inhibitors/Activators | Okadaic acid (PP2A inhibitor); BAPTA-AM (Ca2+ chelator) | Perturb phosphorylation status or signaling context | Pathway manipulation; assessing phosphorylation dynamics [26] [30] |
Biological Context and Pathophysiological Relevance
NDR Kinases in Cellular Morphogenesis and Neuronal Regulation
The functional significance of HM-mediated NDR kinase regulation is exemplified in neuronal development and remodeling processes. In C. elegans, the NDR kinase homolog SAX-1 controls dendrite branch-specific elimination during stress-induced neuronal remodeling, functioning with its conserved interactors SAX-2/Furry and MOB-2 [17]. This system reveals unexpected specificity in pruning processes, with distinct genetic requirements for eliminating different dendritic branch orders.
The SAX-1/NDR pathway promotes endocytosis during neuronal remodeling through functional interactions with the guanine-nucleotide exchange factor RABI-1/Rabin8 and the small GTPase RAB-11.2, linking HM-mediated kinase regulation to membrane dynamics [17]. This illustrates how the fundamental regulatory mechanism of NDR kinases interfaces with specific cellular processes through specialized effector systems.
Hippo Pathway Integration and Therapeutic Implications
NDR/LATS kinases function as essential components of evolutionarily conserved Hippo signaling pathways that control cell proliferation and morphogenesis [1] [2]. In both mammalian cells and model organisms, these kinases form central regulatory nodes that integrate signals from upstream Ste20-family kinases (MST/hippo) and transmit them to diverse downstream effectors.
The critical role of kinase regulation in disease pathophysiology is highlighted by AKT, where HM phosphorylation at Ser473 by mTORC2 is essential for full kinase activation and promotes cell survival and proliferation [27]. Dysregulation of this regulatory mechanism contributes to various pathological conditions, including cancer, metabolic disorders, and neurological diseases, highlighting the therapeutic potential of targeting HM-mediated regulatory systems.
Diagram 2: NDR Kinase Regulatory Logic. This diagram illustrates the integrated regulatory logic of NDR kinase activation, highlighting the essential contributions of HM phosphorylation, MOB coactivator binding, and activation loop phosphorylation in controlling biological outputs [26] [1] [17].
The hydrophobic motif represents a critical structural and functional element that governs kinase activation through phosphorylation-dependent conformational changes and coactivator interactions. In NDR kinases, the HM serves as an integrated regulatory platform that couples upstream MST3-mediated phosphorylation with MOB coactivator binding to control kinase activity with high specificity. The structural basis for MOB binding specificity provides a molecular explanation for the functional segregation of NDR and LATS kinase pathways despite their close evolutionary relationship.
Future research directions include elucidating the structural dynamics of HM-mediated kinase activation at higher resolution, exploring the therapeutic potential of targeting HM regulatory interfaces in disease contexts, and investigating how HM phosphorylation is integrated with subcellular localization and scaffold interactions. The continued mechanistic analysis of HM function will enhance our understanding of kinase regulatory diversity and inform the development of targeted therapeutic strategies for diseases characterized by kinase pathway dysregulation.
The Mps one binder (MOB) family of proteins represents a highly conserved group of signal transducers that play critical roles as kinase activators and adaptors in essential intracellular signaling pathways [15]. First discovered in yeast, MOB proteins are found in all eukaryotes and function primarily through regulatory interactions with serine/threonine kinases of the NDR/LATS family [4] [15]. In mammals, the MOB family has expanded to include multiple members, with MOB1, MOB2, and MOB3 representing distinct classes that exhibit both overlapping and unique functions [15]. This review provides a comprehensive analysis of these three MOB protein classes, with particular emphasis on the structural and mechanistic basis for MOB1 versus MOB2 binding specificity for NDR kinases—a key regulatory node in Hippo and Hippo-like signaling pathways that control cell proliferation, morphogenesis, and genome stability [1] [4] [15].
MOB Family Classification and Structural Characteristics
MOB proteins share a conserved globular fold but have diverged into distinct functional classes through evolution. In animals, MOBs cluster into four classes (I-IV), with MOB1, MOB2, and MOB3 representing Classes I, II, and III, respectively [15].
Table 1: Classification and Key Characteristics of MOB Family Proteins
| Feature | MOB1 (Class I) | MOB2 (Class II) | MOB3 (Class III) |
|---|---|---|---|
| Key Binding Partners | LATS1/2, NDR1/2, MST1/2 | NDR1/2, RAD50 | MST1 (not NDR/LATS) |
| NDR Kinase Activation | Strong activator | Context-dependent (inhibitory or weak activator) | No known interaction |
| Specificity Determinants | Phosphorylation-dependent switch; recognizes HM motif in NDR/LATS | Discrete specificity sites in NTR; competes with MOB1 for NDR binding | Distinct binding surface |
| Cellular Functions | Hippo signaling core component; mitotic exit | Cell cycle progression; DNA damage response; cell motility | Apoptosis regulation; STRIPAK complex component |
| Structural Features | Conserved Mob family fold; phosphoregulation of binding surfaces | Similar globular core but distinct surface properties | Divergent surface characteristics |
The MOB proteins adopt a conserved globular fold with a core consisting of a four alpha-helix bundle, referred to as the "Mob family fold" [15]. Despite this structural conservation, different MOB classes have distinct interaction surfaces that determine their binding specificities and functional outcomes.
MOB1: The Core Hippo Pathway Component
Structure and Activation Mechanism
MOB1 functions as a multifunctional adaptor protein best characterized for its integrative role in regulating Hippo signaling in metazoans and the Mitotic Exit Network in yeast [20]. Human MOB1 binds both upstream kinases (MST1 and MST2) and downstream AGC group kinases (LATS1, LATS2, NDR1, and NDR2) [20]. The binding of MOB1 to MST1 and MST2 is mediated by its phosphopeptide-binding infrastructure, while interaction with LATS and NDR kinases occurs through a distinct interaction surface on MOB1 [20].
A key regulatory mechanism of MOB1 involves phosphorylation-dependent switching between interaction partners. Phosphorylation of MOB1 by MST1/2 enhances its binding to LATS/NDR kinases while reducing affinity for MST kinases themselves, effectively promoting the transfer of MOB1 from upstream to downstream kinases in the pathway [20].
Biological Functions and Pathway Integration
MOB1 serves as an essential core component of the Hippo signaling pathway, which restricts tissue growth and controls organ size in animals [15]. Through its interaction with LATS kinases, MOB1 facilitates phosphorylation and inhibition of YAP/TAZ transcriptional coactivators, thereby repressing genes that promote cell cycle entry and apoptosis resistance [1]. In the context of NDR kinases, MOB1 binding activates these kinases, influencing processes such as cell cycle progression, the DNA damage response, and morphogenesis [4] [13].
MOB2: The NDR-Specific Regulator
Binding Specificity for NDR Kinases
MOB2 exhibits specific binding preference for NDR kinases over LATS kinases, a selectivity that is enforced by discrete structural elements rather than broadly distributed differences [1]. Structural studies of Saccharomyces cerevisiae Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes have revealed that the Ndr/LatsNTR–Mob interface provides a distinctive kinase regulation mechanism [1]. In this mechanism, the Mob cofactor organizes the NDR/LATS N-terminal regulatory (NTR) region to interact with the AGC kinase C-terminal hydrophobic motif (HM), which is involved in allosteric regulation [1].
The specificity of MOB2 for NDR kinases is determined by a short motif in the Mob structure that differs between Mob1 and Mob2 [1]. Alteration of residues in the Cbk1 NTR allows association of the noncognate Mob cofactor, confirming that cofactor specificity is restricted by discrete sites [1].
Functional Diversity of MOB2
MOB2 has been implicated in diverse cellular processes, often with effects opposite to those of MOB1:
Cell Cycle and DNA Damage Response: MOB2 is required to prevent accumulation of endogenous DNA damage and undesired activation of cell cycle checkpoints [4]. MOB2 knockdown triggers a p53/p21-dependent G1/S cell cycle arrest, indicating its role in maintaining normal cell cycle progression [4].
Kinase Regulation: MOB2 competes with MOB1 for NDR binding, with the MOB1/NDR complex corresponding to increased NDR kinase activity and the MOB2/NDR complex being associated with diminished NDR activity [4] [23]. This competition creates a regulatory switch controlling NDR kinase output.
Cancer Cell Motility: In hepatocellular carcinoma cells, MOB2 knockout promotes migration and invasion, while MOB2 overexpression has the opposite effect [23]. Mechanistically, MOB2 regulates the alternative interaction of MOB1 with NDR1/2 and LATS1, leading to increased phosphorylation of LATS1 and MOB1 and consequent inactivation of YAP [23].
MOB3: The Distinctive Family Member
Unique Binding Properties
MOB3 proteins (including MOB3A, MOB3B, and MOB3C) represent the most divergent class of MOB family proteins. Unlike MOB1 and MOB2, MOB3 proteins do not form complexes with NDR or LATS kinases [4]. Instead, MOB3 associates with the pro-apoptotic kinase MST1 (STK4), indicating a distinct functional role separate from the core NDR/LATS signaling axis [4].
Emerging Functional Roles
While less characterized than MOB1 and MOB2, emerging evidence suggests MOB3 proteins function as components of the STRIPAK complex (Striatin Interacting Phosphatase and Kinase), which antagonizes Hippo signaling [15]. This places MOB3 in an opposing regulatory position to MOB1, potentially creating a balance system for fine-tuning pathway output.
Structural Basis of MOB-NDR/LATS Interactions
Molecular Determinants of Binding Specificity
Structural studies have revealed the detailed molecular interactions governing MOB-kinase specificity. The improved crystal structure of the Cbk1NTR–Mob2 complex shows that the NTR forms a bihelical conformation similar to that observed in LatsNTR– and NdrNTR–Mob1 structures [1]. This NTR region binds to Mob coactivator proteins to form complexes that are essential and evolutionarily conserved components of "Hippo" signaling pathways [1].
The interface between MOB proteins and their kinase partners serves as a structural platform that mediates kinase-cofactor binding and organizes the kinase regulatory domains. The Mob-organized NTR appears to mediate association of the HM with an allosteric site on the N-terminal kinase lobe, positioning the phosphorylated HM for optimal kinase activation [1].
Comparative Analysis of MOB1 vs. MOB2 Binding to NDR Kinases
Table 2: Quantitative Comparison of MOB1 and MOB2 Binding to NDR Kinases
| Parameter | MOB1-NDR Complex | MOB2-NDR Complex |
|---|---|---|
| Binding Affinity | High (enhanced by phosphorylation) | Context-dependent |
| Kinase Activation | Strong activation | Inhibition or weak activation |
| Cellular Outcome | Enhanced NDR signaling | Attenuated NDR signaling |
| Competitive Relationship | Displaced by MOB2 overexpression | Competes with MOB1 for NDR binding |
| Structural Features | Phospho-dependent conformational switch | Discrete specificity determinants in NTR |
The competition between MOB1 and MOB2 for NDR binding creates a regulatory switch that controls NDR kinase activity [4] [23]. This competition is particularly significant given that MOB2 expression can block MOB1-mediated NDR activation, potentially redirecting signaling outcomes [4].
Experimental Approaches for Studying MOB-Kinase Interactions
Structural Biology Techniques
Understanding MOB-kinase interactions has relied heavily on structural biology approaches:
X-ray crystallography has provided high-resolution structures of MOB-kinase complexes, such as the Cbk1NTR–Mob2 and Dbf2NTR–Mob1 structures solved at 2.8 Å and 3.5 Å resolution, respectively [1].
Cryo-electron microscopy (cryo-EM) enables determination of structural transitions of proteins in native cell membranes and entire cells [31].
Nuclear Magnetic Resonance (NMR) spectroscopy delivers detailed insights into small molecule and protein dynamics in solution, useful for fragment-based screening and conformational analysis [32].
Biochemical and Cellular Assays
Functional characterization of MOB proteins employs diverse experimental approaches:
Kinase activity assays to measure MOB-dependent activation of NDR/LATS kinases [13]
Co-immunoprecipitation and pull-down assays to assess protein-protein interactions and competition between MOB1 and MOB2 [23]
Cellular localization studies using fluorescently tagged proteins to examine subcellular distribution and co-localization [13]
Genetic manipulation including knockout, knockdown, and overexpression to determine functional consequences [23]
Research Reagent Solutions
Table 3: Essential Research Tools for MOB Protein Studies
| Reagent/Tool | Function/Application | Examples/Notes |
|---|---|---|
| Crystallization Constructs | Structural studies of MOB-kinase complexes | Engineered zinc-binding Mob2 (V148C Y153C) for improved stability [1] |
| Phospho-specific Antibodies | Detection of activation-specific phosphorylation | Anti-NDR pThr444, Anti-NDR pSer281 [13] |
| Inducible Expression Systems | Controlled MOB expression for functional studies | Inducible membrane-targeted hMOB1A for studying rapid NDR activation [13] |
| CRISPR/Cas9 Tools | Gene knockout and editing | sgRNA targeting MOB2: 5'-AGAAGCCCGCTGCGGAGGAG-3' [23] |
| Lentiviral Vectors | Stable gene expression or knockdown | LV-MOB2 for overexpression; lentiCRISPRv2 for knockout [23] |
Signaling Pathway Diagrams
MOB Protein Signaling Network
This diagram illustrates the complex regulatory network involving MOB proteins. MOB1 (green) serves as a core activator of both LATS and NDR kinases downstream of MST1/2 kinases. MOB2 (red) competes with MOB1 for NDR binding, creating a regulatory switch. MOB3 (blue) functions as part of the STRIPAK complex that negatively regulates MST kinases. The integration of these opposing functions allows fine-tuning of pathway output, particularly the phosphorylation status of YAP and its subsequent transcriptional activity.
MOB-NDR Binding Specificity and Outcomes
This diagram illustrates the competitive binding of MOB1 and MOB2 to NDR kinases and the divergent functional outcomes. MOB1 binding activates NDR kinase, promoting signaling outputs such as proper cell cycle progression and DNA damage response. In contrast, MOB2 binding results in a low-activity NDR complex, altering signaling output and potentially redirecting cellular responses. The competitive relationship between MOB1 and MOB2 creates a regulatory switchpoint for controlling NDR kinase activity.
The comparative analysis of MOB family proteins reveals a sophisticated regulatory network centered on NDR/LATS kinases. MOB1 and MOB2, despite their structural similarities, have evolved distinct binding specificities and functional outcomes through discrete structural determinants. MOB1 serves as a strong activator of both LATS and NDR kinases, positioning it as a core component of Hippo signaling. In contrast, MOB2 exhibits specific binding to NDR kinases with context-dependent inhibitory effects, creating a competitive regulatory switch with MOB1. MOB3 represents a more divergent functional branch, associating with alternative partners like MST1 and the STRIPAK complex.
The structural basis for MOB1 versus MOB2 binding specificity for NDR kinases, governed by discrete sites in the NTR region rather than broadly distributed differences, provides a paradigm for understanding how homologous regulatory proteins achieve functional diversity [1]. This specificity has profound biological implications, as the competition between MOB1 and MOB2 for NDR binding creates a tunable switch that can redirect signaling outcomes in processes ranging from cell cycle control to cell motility [4] [23].
Future research should focus on elucidating the precise structural transitions that occur upon MOB-kinase binding, the spatial and temporal regulation of MOB expression and localization, and the potential therapeutic targeting of these interactions in diseases such as cancer. The emerging role of MOB proteins as integrators of multiple signaling inputs positions them as key nodes for therapeutic intervention in pathways controlling cell growth and survival.
Experimental Approaches for Characterizing MOB-NDR Interactions and Cellular Functions
Crystallography and Cryo-EM Techniques for Structural Determination
A comprehensive understanding of cellular signaling pathways requires precise structural knowledge of their core components. The Hippo signaling pathway, an evolutionarily conserved system controlling cell proliferation and morphogenesis, relies on the specific partnership between NDR/LATS kinases and Mob coactivator proteins [1] [2]. Within this family, a striking specificity exists: LATS kinases associate exclusively with Mob1 proteins, while NDR kinases bind specifically to Mob2 proteins [1]. This selective binding is crucial for proper pathway function, with misregulation linked to carcinogenesis [8]. Determining the three-dimensional structures of these complexes is therefore essential not only for understanding fundamental biology but also for targeted drug development.
Two primary techniques—X-ray crystallography and cryo-electron microscopy (cryo-EM)—have enabled researchers to visualize these interactions at atomic resolution. This technical guide examines both methods within the context of Mob-NDR kinase research, detailing their respective workflows, capabilities, and applications for elucidating the structural basis of binding specificity. We will explore how these techniques have revealed the novel kinase-coactivator organization and autoinhibitory mechanisms that govern Hippo pathway signaling, providing a framework for selecting appropriate structural approaches for specific research objectives in kinase-coactivator systems.
Core Structural Techniques: Principles and Applications
X-ray Crystallography
X-ray crystallography has been the workhorse technique for determining high-resolution structures of kinase-coactivator complexes. This method involves purifying the protein complex, forming ordered crystals, collecting X-ray diffraction data, and solving the structure through molecular replacement or other phasing methods.
Key Applications in Mob-NDR Research:
- Determining the first structure of the Cbk1NTR–Mob2 complex from Saccharomyces cerevisiae at 2.8 Å resolution [1]
- Revealing the autoinhibited structure of full-length MOB1B, showing how its N-terminal extension forms a β-strand (SN strand) and a Switch α-helix that blocks the LATS1-binding surface [33]
- Elucidating the specific binding interfaces between NDR/LATS kinases and their cognate Mob cofactors, highlighting discrete residues that enforce binding specificity rather than broadly distributed regions [1]
Cryo-Electron Microscopy (Cryo-EM)
Cryo-EM has emerged as a powerful complementary technique that can resolve structures without crystallization. This method involves flash-freezing protein samples in vitreous ice, collecting images of individual particles, and computationally reconstructing three-dimensional structures.
Technical Advances and Capabilities:
- Direct electron detectors have dramatically improved resolution by enabling movie mode acquisition that corrects for specimen movement [34]
- Volta phase plates can enhance contrast for small proteins, though they require precise focusing [35]
- Scaffold-based approaches (e.g., DARPin cages, coiled-coil fusions) enable structure determination of small proteins like kRasG12C (19 kDa) by increasing effective particle size [35]
Table 1: Comparison of Structural Techniques for Kinase-Cofactor Complexes
| Parameter | X-ray Crystallography | Single-Particle Cryo-EM |
|---|---|---|
| Typical Sample Requirement | High purity, crystallizable | High purity, >50 kDa (native or scaffold-fused) |
| Resolution Range | 1.5-3.5 Å (for successful complexes) | 3.0-4.5 Å (challenging for small complexes) |
| Key Advantage | High resolution, well-established | Bypasses crystallization, captures dynamic states |
| Mob-NDR Application Example | Cbk1NTR–Mob2 (2.8 Å) [1] | kRasG12C-APH2 scaffold (3.7 Å) [35] |
| Limitations | Requires crystallization, static snapshot | Lower resolution for small targets, complex data processing |
Structural Insights into MOB-NDR Kinase Specificity
Determinants of Binding Specificity
Structural studies have revealed that Mob coactivators bind to the characteristic N-terminal regulatory (NTR) region of NDR/LATS kinases [1]. The NTR forms a V-shaped helical hairpin that interfaces with the Mob protein surface. Comparative analysis of Cbk1NTR-Mob2 and Dbf2NTR-Mob1 structures indicates that cofactor specificity is restricted by discrete sites rather than being broadly distributed across the interface [1]. Mutation of these specific residues allows association with noncognate Mob cofactors, demonstrating the structural basis for the observed binding preferences in physiological contexts.
Autoinhibition and Activation Mechanisms
The structure of full-length MOB1B revealed a key autoinhibition mechanism, with the N-terminal extension forming a Switch helix that blocks the LATS1-binding surface [33]. This autoinhibition is stabilized by β-sheet formation between the SN strand of the N-terminal extension and the S2 strand of the MOB1 core domain. Phosphorylation of Thr12 and Thr35 residues by upstream kinases structurally accelerates dissociation of the Switch helix through a "pull-the-string" mechanism, enabling LATS1 binding and kinase activation [33].
Functional Consequences for Kinase Regulation
The Mob-organized NTR appears to mediate association of the C-terminal hydrophobic motif (HM) with an allosteric site on the N-terminal kinase lobe [1]. This configuration provides a distinctive kinase regulation mechanism where Mob binding positions the phosphorylated HM (e.g., Thr-743 in Cbk1) proximal to a conserved Arg in the NTR, facilitating optimal orientation of the kinase's αC helix, a critical component for kinase activation [1] [2].
Diagram 1: MOB Cofactor Specificity in Hippo Signaling. The diagram illustrates the specific binding partnerships between MOB cofactors and their cognate NDR/LATS kinases, leading to distinct cellular outcomes. This specificity is structurally enforced by discrete interfacial residues.
Experimental Protocols for Structural Studies
Crystallography Workflow for Kinase-Cofactor Complexes
Sample Preparation and Crystallization:
- Construct Design: Express kinase NTR domains (e.g., Cbk1 residues 251-351) and Mob cofactors (e.g., Mob2 45-287) in E. coli. Engineering stabilizing mutations (e.g., Mob2 V148C Y153C) can improve expression and crystallization [1].
- Complex Formation: Purify individual components and mix at equimolar ratios. Isolate the complex using size-exclusion chromatography.
- Crystallization: Screen commercial crystallization kits using robotic liquid handlers. Optimize initial hits by varying pH, precipitant concentration, and temperature.
- Cryoprotection: Transfer crystals to cryoprotectant solutions (e.g., 25% glycerol) before flash-freezing in liquid nitrogen.
Data Collection and Structure Determination:
- X-ray Data Collection: Collect diffraction data at synchrotron beamlines (e.g., wavelength ~0.97872Å). Monitor for radiation damage.
- Structure Solution: Use molecular replacement with known Mob or kinase NTR structures as search models (e.g., PDB 4LQS for Cbk1-Mob2) [2].
- Model Building and Refinement: Iteratively build the model using Coot and refine with Phenix or REFMAC. Validate geometry using MolProbity.
Table 2: Crystallographic Data Collection and Refinement Statistics for NDR/LATS-Mob Complexes
| Parameter | Cbk1NTR–Mob2 | Dbf2NTR–Mob1 | Cbk1–Mob2–pepSsd1 |
|---|---|---|---|
| Resolution (Å) | 2.8 | 3.5 | 3.15 |
| Space Group | P4₁2₁2 | P6₁22 | C121 |
| Rwork/Rfree | 0.249/0.284 | 0.229/0.263 | 0.231/0.298 |
| Bond Lengths RMSD (Å) | 0.004 | 0.010 | 0.011 |
| Bond Angles RMSD (°) | 0.70 | 0.75 | 1.35 |
| PDB Accession | To be deposited | To be deposited | To be deposited |
Cryo-EM Workflow for Small Protein Complexes
Sample Preparation and Grid Preparation:
- Scaffold Design for Small Targets: For proteins <50 kDa, fuse to stabilizing scaffolds. For kRasG12C (19 kDa), fuse C-terminal helix to APH2 coiled-coil motif recognized by nanobodies [35].
- Vitrification: Apply 3-4 μL sample to glow-discharged EM grids. Blot to thin layer (∼20-300 nm) and plunge-freeze in liquid ethane.
- Quality Assessment: Screen grids for ice quality and particle distribution using screening microscope.
Data Collection and Processing:
- Imaging Parameters: Collect data using 200-300 keV microscope with direct electron detector. Use defocus range of -0.5 to -2.5 μm. Limit electron dose to 10-20 e⁻/Ų to minimize radiation damage [34].
- Movie Processing: Correct for beam-induced motion and dose-weight frames using MotionCor2 or similar.
- Particle Picking and Classification: Use template-based or neural net picking. Perform 2D and 3D classification to isolate homogeneous particles.
- Map Reconstruction and Model Building: Reconstruct initial map, refine iteratively, and build atomic model using Coot or ISOLDE.
Diagram 2: Structural Biology Workflow Comparison. The diagram outlines parallel workflows for cryo-EM and X-ray crystallography, highlighting key steps from sample preparation to structure deposition. Scaffold fusion (cryo-EM) and complex formation (crystallography) represent critical preparatory steps specific to each method.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagents for MOB-NDR Structural Studies
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Expression Systems | E. coli BL21(DE3), Bac-to-Bac Baculovirus | Recombinant protein expression for crystallography and cryo-EM samples |
| Crystallization Screens | Hampton Research Crystal Screens, MemGold | Initial crystallization condition identification |
| Cryo-EM Scaffolds | APH2 coiled-coil, DARPin cages, HR00C3_2 | Size enhancement for small protein targets (<50 kDa) |
| Detection Reagents | Anti-His antibodies, Streptavidin gold conjugates | Sample quality verification and cryo-EM fiducial markers |
| Stabilizing Mutations | Mob2 V148C Y153C (zinc-binding) | Improved protein stability and crystallization propensity |
| Data Collection Facilities | Synchrotron beamlines (e.g., APS, ESRF), Titan Krios microscopes | High-resolution data collection for crystallography and cryo-EM |
The complementary application of X-ray crystallography and cryo-EM has dramatically advanced our understanding of MOB-NDR kinase specificity and regulation. Crystallography has provided atomic-resolution snapshots of specific complexes like Cbk1-Mob2, revealing the precise interfacial residues that determine binding specificity [1]. Meanwhile, cryo-EM offers growing potential for studying these complexes in more native states and for challenging targets that resist crystallization [35].
For researchers investigating MOB1 versus MOB2 binding specificity, crystallography remains ideal for characterizing well-behaved complexes and mapping precise interaction interfaces. Cryo-EM shows particular promise for studying full-length kinases in different activation states, complexes with scaffolding proteins, and the structural effects of cancer-associated mutations in NDR2 [8]. As cryo-EM methodologies continue advancing, particularly for smaller proteins, this technique will likely play an increasingly central role in elucidating the structural nuances of Hippo signaling and developing therapeutic strategies that target these specific interactions.
Site-Directed Mutagenesis to Probe Specificity Determinants
The Mps One Binder (MOB) proteins are evolutionarily conserved eukaryotic signal transducers that play pivotal roles as regulators of NDR/LATS kinases in Hippo signaling pathways. A central question in this field concerns the molecular mechanisms that dictate the binding specificity of MOB1 versus MOB2 for their NDR kinase partners (NDR1/STK38 and NDR2/STK38L). This specificity is biologically crucial, as MOB1 typically functions as a kinase activator, while MOB2 can act as a negative regulator that competes with MOB1 for NDR binding [4]. Understanding these determinants is essential for deciphering Hippo pathway regulation and developing targeted therapeutic interventions. This technical guide provides a comprehensive framework for employing site-directed mutagenesis to probe the specificity determinants governing MOB1 versus MOB2 binding specificity for NDR kinases, presenting detailed methodologies, structural insights, and experimental protocols for researchers investigating this critical signaling axis.
Structural Foundations of MOB-NDR Interactions
The structural basis for MOB-NDR interactions has been elucidated through crystallographic studies of complexes such as MOB1-NDR2 and Cbk1-Mob2. MOB proteins adopt a globular structure composed of a conserved four-helix bundle stabilized by a zinc atom [36] [22]. The N-terminal regulatory domain (NTR) of NDR kinases, comprising approximately 70 amino acids, forms a V-shaped structure of two antiparallel α-helices that engages with the MOB protein surface [22].
The binding interface is characterized by complementary electrostatic interactions: a conserved, negatively charged surface on MOB proteins interacts with positively charged residues in the NTR of NDR kinases [22]. This interaction mode is evolutionarily conserved from yeast to humans, as demonstrated by the structure of the yeast Cbk1-Mob2 complex, which revealed a novel coactivator-organized activation region unique to NDR/LATS kinases [2] [14].
Key Specificity Determinants in MOB Proteins
Structural comparisons between MOB1-NDR2 and MOB1-LATS1 complexes have identified crucial residues that dictate binding specificity. Asp63 in MOB1 emerges as a critical specificity determinant—it forms a specific bond with His646 in LATS1 but does not participate in NDR2 binding [22]. This key difference enables the design of mutagenesis strategies to dissect functional contributions.
Table 1: Key Specificity Determinants in MOB Proteins
| Residue | Location | Function | Effect of Mutation |
|---|---|---|---|
| Asp63 | MOB1 | Specific interaction with LATS1/2 His646 | Disrupts MOB1-LATS binding while preserving MOB1-NDR binding [22] |
| Cluster of surrounding residues (Phe642, Met643, Gln645, Val647, Val650) | LATS1 | Supports His646 interaction with MOB1 Asp63 | Combined mutations enhance binding disruption [22] |
| Phe31 | NDR2 | Does not interact with MOB1 Asp63 | NDR binding to MOB1 is unaffected by MOB1 Asp63 mutations [22] |
Critical Interaction Residues in NDR Kinases
The N-terminal regulatory domain of NDR kinases contains highly conserved positively charged residues that form specific contacts with MOB proteins. Structural and biochemical studies have validated the importance of these residues through alanine scanning mutagenesis.
Table 2: Critical NDR Residues for MOB Binding
| NDR2 Residue | MOB1 Interaction Partner | Interaction Type | Functional Validation |
|---|---|---|---|
| Lys25 | Leu36, Gly39, Leu41, Ala44, Gln67, Met70, Leu71, Leu173, Gln174, His185 of MOB1 | Hydrogen bonds, van der Waals | K24A (NDR1) reduces MOB binding and kinase activation [22] |
| Tyr32 | Same as above | Hydrogen bonds, van der Waals | Y31A (NDR1) impairs MOB binding [22] |
| Arg42 | Glu51, Glu55, Trp56, Val59, Phe132, Pro133, Lys135, Val138 of MOB1 | Hydrogen bonds, van der Waals | R41A (NDR1) reduces MOB binding and kinase activation [22] |
| Arg45 | Intramolecular with Glu74 | Stabilizes V-shape of NTR | R44A (NDR1) impairs MOB binding [22] |
| Arg79 | Same as Arg42 | Hydrogen bonds, van der Waals | R78A (NDR1) reduces MOB binding and kinase activation [22] |
Figure 1: MOB-NDR/LATS Interaction Network and Specificity Determinants. MOB1 activates both NDR and LATS kinases, while MOB2 competes with MOB1 for NDR binding. The Asp63-His646 interaction specifically mediates MOB1-LATS binding.
Experimental Design and Mutagenesis Strategies
Rational Mutagenesis Design Principles
When designing mutagenesis experiments to probe MOB-NDR specificity, consider these key structural principles:
Electrostatic Complementarity: Target residues forming charged interactions at the binding interface, particularly the conserved negative surface on MOB proteins and positive patches on NDR N-terminal domains [22].
Conservation Analysis: Prioritize residues conserved across orthologs but divergent between paralogs, as these often determine functional specificity [37].
Structural Clustering: Focus on residues that cluster in three-dimensional space despite linear sequence separation, as these often form functional epitopes [37] [22].
Functional Validation: Design mutations that test specific mechanistic hypotheses (e.g., charge-reversal versus alanine substitutions to distinguish electrostatic versus steric effects).
Key Mutagenesis Targets for Specificity Profiling
Based on structural data, the following residues represent high-value targets for probing MOB-NDR specificity:
MOB1-Specific Targets:
- Asp63: Create D63A (loss-of-function) and D63K (charge-reversal) mutants to selectively disrupt MOB1-LATS binding while preserving MOB1-NDR interactions [22].
- Phosphoregulation sites: Thr12 and Thr35, which are phosphorylated by MST1/2, regulate MOB1 conformation and binding accessibility [38].
MOB2-Specific Targets:
- Equivalent specificity residues: Identify and mutate residues that may confer MOB2's preferential binding to NDR over LATS kinases.
- Negative regulatory surface: Map regions that mediate MOB2's ability to compete with MOB1 and potentially inhibit NDR activation [4].
NDR Kinase Targets:
- NTR basic residues: Lys25, Arg42, Arg45, and Arg79 in NDR2 are critical for MOB binding [22].
- Hydrophobic motif: Thr442 in NDR2 (equivalent to Thr444 in NDR1) is phosphorylated by upstream kinases like MST3 and is essential for full kinase activation [26].
Experimental Protocols and Methodologies
Site-Directed Mutagenesis Protocol
This optimized protocol enables efficient introduction of point mutations into MOB and NDR constructs for specificity studies:
Materials:
- High-fidelity DNA polymerase (e.g., PfuUltra)
- DpnI restriction enzyme
- Competent E. coli cells (XL1-Blue recommended)
- Plasmid DNA templates for MOB1, MOB2, NDR1, and NDR2
- Qiafilter Maxiprep kit (QIAGEN) for plasmid purification [26]
Method:
- Primer Design: Design complementary primers containing desired mutations with 15-20 bp flanking sequences on each side. For MOB1 Asp63 mutants:
- Forward: 5'-CACCTAC[G→C]TTCGACCC-3' (example for D63A)
- Annealing temperature: ~60°C
PCR Amplification:
- 30 ng template DNA
- 125 ng each primer
- 200 μM dNTPs
- 2.5 U PfuUltra in 1× buffer
- Cycling: 95°C 2 min; [95°C 30s, 60°C 1min, 68°C 6min] × 18 cycles; 68°C 10min
Template Digestion: Add 1 μL DpnI directly to PCR reaction, incubate at 37°C for 1 hour to digest methylated parental DNA.
Transformation: Transform 2 μL reaction into 50 μL competent XL1-Blue cells, plate on selective media.
Sequence Verification: Isolate plasmids from resulting colonies and verify mutations by Sanger sequencing using appropriate sequencing primers [26].
Binding Affinity Measurement Using Fluorescence Polarization
Quantify the impact of mutations on MOB-NDR binding affinity using fluorescence polarization:
Reagent Preparation:
- Express and purify recombinant wild-type and mutant MOB and NDR proteins as N-terminal dual 6xhistidine-glutathione S-transferase (GST) TEV-cleavable fusion proteins using modified pETM-30 vector in E. coli BL21(DE3) CodonPlus RIL cells [38].
Purify MOB proteins in batch on glutathione-Sepharose resin, elute by cleavage from affinity tags with HIS-tagged TEV protease.
Remove TEV protease by subtractive immobilized-metal affinity chromatography.
Concentrate and buffer-exchange cleaved protein by size exclusion chromatography using Superdex 75 column [38].
Label NTR peptides of NDR kinases with fluorescent dyes (e.g., FITC) for polarization assays.
Binding Assay:
- Prepare serial dilutions of MOB proteins (0.1 nM - 10 μM) in assay buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl).
Mix constant concentration of fluorescent NTR peptide (1 nM) with MOB protein dilutions.
Incubate 30 minutes at room temperature in dark.
Measure fluorescence polarization using plate reader with appropriate filters.
Fit data to single-site binding model to calculate dissociation constants (Kd).
Compare Kd values between wild-type and mutant proteins to quantify binding energy changes [38] [2].
Functional Kinase Activation Assays
Determine how MOB mutations affect NDR kinase activity through in vitro kinase assays:
Kinase Reaction Setup:
- Immunoprecipitation: Express HA-tagged NDR2 wild-type and mutants in COS-7 or HEK293F cells. Lyse cells in IP buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 10% glycerol, protease and phosphatase inhibitors). Immunoprecipitate using anti-HA antibodies [26].
Kinase Reaction:
- Resuspend immunoprecipitates in 30 μL kinase buffer (20 mM HEPES, pH 7.4, 10 mM MgCl₂, 1 mM DTT)
- Add 100 μM ATP, 1 μCi [γ-³²P]ATP, and appropriate substrate (e.g., 1 μg myelin basic protein)
- Incubate 30 minutes at 30°C
- Stop reaction with SDS sample buffer
Analysis: Separate proteins by SDS-PAGE, transfer to PVDF membrane, visualize phosphorylation by autoradiography, quantify using phosphoimager [26].
Activation-Specific Phosphorylation: Monitor phosphorylation of activation sites (Ser281/Ser282 in activation loop and Thr444/Thr442 in hydrophobic motif) using phospho-specific antibodies [26].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for MOB-NDR Specificity Studies
| Reagent Category | Specific Examples | Function/Application | Source/Reference |
|---|---|---|---|
| Expression Plasmids | pCMV5-HA-NDR2, pCMV5-HA-MST3, myc-C1-MOB1A, pEBG2T-MST3, pTER-shMST3 | Mammalian expression and knockdown | [26] |
| Bacterial Expression Vectors | pETM-30 (modified), pGEX-2T-MOB1A | Recombinant protein production | [26] [38] |
| Cell Lines | HEK293F, COS-7 | Transfection, immunoprecipitation, kinase assays | [26] |
| Antibodies | Anti-P-Ser282, Anti-P-Thr442 (NDR), 12CA5 (HA), 9E10 (myc), anti-GST, anti-NDR-NT peptide antibody | Detection, immunoprecipitation, phospho-specific monitoring | [26] |
| Purification Tools | Glutathione-Sepharose resin, HIS-tagged TEV protease, Superdex 75 SEC columns | Protein purification and cleavage | [38] |
| Kinase Assay Components | [γ-³²P]ATP, myelin basic protein, kinase buffer | In vitro kinase activity measurement | [26] |
Figure 2: Experimental Workflow for Probing MOB-NDR Specificity Determinants. The systematic approach from structural analysis to functional validation ensures comprehensive characterization of specificity determinants. Key considerations at each stage enhance experimental robustness.
Data Interpretation and Validation
Analyzing Mutational Impact on Binding Energetics
When interpreting mutagenesis results, consider these quantitative frameworks:
Binding Affinity Changes: Calculate ΔΔG = RTln(Kdmutant/KdWT) to determine the energetic contribution of specific residues.
Specificity Indices: Compute specificity ratios (KdMOB1-mutant/KdMOB2-wt) / (KdMOB1-wt/KdMOB2-wt) to quantify how mutations alter MOB1 versus MOB2 preference.
Structural Correlations: Map binding energy changes onto structural models to distinguish direct binding effects from conformational perturbations.
Functional Validation in Cellular Contexts
Translate in vitro findings to cellular systems using these approaches:
Co-immunoprecipitation: Express wild-type and mutant MOB/NDR constructs in HEK293F cells, immunoprecipitate with anti-HA or anti-myc antibodies, and detect binding partners by Western blotting [26].
Kinase-Dead Mutants: Utilize kinase-dead MST3 (MST3KR) to probe upstream regulation of NDR hydrophobic motif phosphorylation [26].
Knockdown/Rescue Experiments: Deplete endogenous MOB1/2 using shRNA (e.g., pTER-shMST3 vectors) and rescue with RNAi-resistant wild-type or mutant constructs to assess functional complementation [26] [4].
Phenotypic Assays: Monitor downstream processes such as cell cycle progression, dendrite pruning (for neuronal contexts), or transcriptional readouts to connect molecular interactions to biological functions [17] [4].
Site-directed mutagenesis provides a powerful approach for deciphering the molecular determinants that govern MOB1 versus MOB2 binding specificity for NDR kinases. The structural insights, experimental protocols, and analytical frameworks presented in this technical guide establish a comprehensive foundation for designing and interpreting mutagenesis studies in this biologically crucial system. By systematically probing the interface between MOB proteins and NDR kinases, researchers can advance our understanding of Hippo pathway regulation and identify potential therapeutic targets for diseases involving dysregulated cell growth and proliferation. The integration of structural biology with quantitative biochemical assays and functional validation in cellular models represents the most robust approach for establishing definitive mechanisms of specificity in this evolutionarily conserved signaling axis.
Co-immunoprecipitation and Pull-Down Assays for Interaction Validation
The validation of specific protein-protein interactions is a cornerstone of molecular biology, particularly in the study of complex cellular signaling pathways. Within the context of Hippo signaling networks, the specific binding between MOB coactivators and NDR/Lats kinases represents a critical regulatory node controlling fundamental processes including cell proliferation, morphogenesis, and cell division. Co-immunoprecipitation (Co-IP) and pull-down assays have emerged as indispensable techniques for confirming these interactions under conditions that preserve physiological relevance. These complementary approaches allow researchers to capture protein complexes from native cellular environments or reconstructed systems, providing direct evidence of molecular associations that drive cellular function.
The significance of these methods is particularly evident in distinguishing the precise binding preferences between highly homologous protein families, such as the specific association of MOB1 with Lats kinases and MOB2 with Ndr kinases. As research progresses toward therapeutic targeting of these interactions, robust validation assays become increasingly crucial for drug discovery efforts. This technical guide examines the principles, applications, and methodological considerations for employing Co-IP and pull-down assays, with specific emphasis on their role in elucidating MOB-NDR kinase interactions within broader thesis research on Hippo signaling components.
Core Principles and Comparative Analysis
Co-immunoprecipitation (Co-IP) Fundamentals
Co-immunoprecipitation is an antibody-based technique designed to isolate a target protein ("bait") along with its direct and indirect binding partners ("prey") from a complex biological mixture. The fundamental principle relies on the specific interaction between an antibody and its target antigen, which subsequently allows the entire protein complex to be captured onto a solid support, typically beads coated with Protein A, Protein G, or specific binding ligands [39] [40]. When performed under non-denaturing conditions, Co-IP preserves transient protein interactions that occur in physiological contexts, making it particularly valuable for studying signaling complexes such as those formed between MOB proteins and NDR kinases.
The standard Co-IP workflow encompasses three critical stages: sample preparation, immunoprecipitation, and elution/analysis. During sample preparation, cells are lysed using mild, non-denaturing buffers that maintain protein-protein interactions while releasing cellular contents [40]. The lysate is then incubated with a specific antibody against the bait protein, forming antigen-antibody complexes. These complexes are subsequently captured using bead-based supports, washed to remove non-specifically bound proteins, and finally eluted for downstream analysis by western blotting or mass spectrometry [39]. A critical advantage of Co-IP for MOB-NDR interaction studies is its applicability to primary cells and tissues, allowing investigation of these complexes in a natural cellular environment [41].
Pull-Down Assay Fundamentals
Pull-down assays share conceptual similarity with Co-IP in their goal of isolating protein complexes, but differ fundamentally in their capture mechanism. Rather than employing antibodies, pull-down assays utilize a purified, immobilized "bait" protein to capture binding partners ("prey") from a mixture [39] [42]. The bait protein is typically tagged with an affinity epitope (GST, His, FLAG, etc.) and bound to a resin specific for the tag. When a cell lysate or mixture of purified proteins is applied, interacting proteins bind to the immobilized bait and can be recovered after washing steps.
This approach offers distinct advantages for studying MOB-NDR interactions, particularly in allowing precise control over which protein component is used as bait and enabling characterization of direct binary interactions without potential interference from additional cellular components. Common pull-down variations include GST pull-down, His-tag pull-down, and streptavidin-biotin based systems, each with particular strengths for different experimental scenarios [42].
Technique Comparison and Selection Criteria
Table 1: Comparison of Co-IP and Pull-Down Assay Characteristics
| Parameter | Co-Immunoprecipitation | Pull-Down Assay |
|---|---|---|
| Basis of Capture | Antibody-antigen specificity | Affinity tag recognition |
| Interaction Context | Native cellular environment | Defined system (native or reconstituted) |
| Throughput | Moderate | Moderate to high |
| Key Applications | Identification of novel binding partners; Validation of in vivo interactions | Confirmation of direct interactions; Mapping interaction domains |
| Sample Requirements | Cell lysates, tissue extracts | Cell lysates or purified proteins |
| Critical Reagents | Specific, high-quality antibodies | Tagged proteins, affinity resins |
| Advantages | Studies endogenous complexes; Preserves physiological context | No antibody requirement; Controls for direct binding |
| Limitations | Antibody specificity concerns; Cannot distinguish direct/indirect interactions | May miss native context; Requires protein tagging |
The choice between Co-IP and pull-down assays depends heavily on the research question and available reagents. For initial discovery of novel binding partners under physiological conditions, Co-IP is typically preferred. When investigating direct interactions or working with proteins lacking high-quality antibodies, pull-down assays offer a viable alternative [39] [42]. In the context of MOB-NDR kinase research, these techniques are often used complementarily—Co-IP validates interactions in cellular contexts, while pull-down assays confirm direct binding and elucidate structural determinants of specificity.
MOB1 vs. MOB2 Binding Specificity for NDR Kinases
Structural Basis of Specific MOB-NDR/Lats Interactions
The specific binding preferences between MOB coactivators and NDR/Lats kinases represent a paradigm of selective molecular recognition in eukaryotic signaling systems. Structural biology approaches have revealed that Ndr/Lats kinases contain a characteristic N-terminal regulatory (NTR) region that functions as the primary binding site for Mob cofactors, with Lats kinases specifically associating with Mob1 proteins and Ndr kinases with Mob2 proteins [1]. This precise molecular discrimination occurs despite significant sequence conservation across the entire Ndr/Lats kinase family, suggesting evolutionary refinement of interaction specificity.
Crystallographic studies of Saccharomyces cerevisiae Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes have provided high-resolution insight into the molecular determinants of this specificity. The improved structure of the Cbk1–Mob2 complex reveals that the NTR forms a V-shaped helical hairpin that interfaces with the Mob cofactor, creating a structural platform that mediates kinase-cofactor binding [1]. This interface organizes the NDR kinase NTR to interact with the C-terminal hydrophobic motif (HM), a critical element in allosteric kinase regulation. The Mob-cofactor interaction appears to position the HM for optimal engagement with an allosteric site on the N-terminal kinase lobe, thereby facilitating kinase activation.
Determinants of Binding Specificity
Research indicates that binding specificity between MOB proteins and NDR kinases is governed by discrete molecular recognition sites rather than broadly distributed interface properties. In Saccharomyces cerevisiae, the Ndr-family kinase Cbk1 associates specifically with Mob2, while the Lats-related kinases Dbf2 and Dbf20 bind exclusively to Mob1, despite simultaneous presence of all proteins in the cytosol [1]. This specificity is maintained through a short motif in the Mob structure that differs between Mob1 and Mob2, with alteration of key residues in the Cbk1 NTR enabling association with noncognate Mob cofactors.
In human systems, similar specificity principles operate, with NDR1 and NDR2 kinases forming stable complexes with human MOB2 but not MOB1, despite structural similarities [3]. This association dramatically stimulates NDR1 and NDR2 catalytic activity, identifying MOB proteins as a unique class of kinase-activating subunits that may be functionally analogous to cyclins. The differential subcellular localization of NDR1 (nuclear) and NDR2 (punctate cytoplasmic) further suggests that each kinase may serve distinct functions despite similar MOB binding preferences [3].
Table 2: Key Research Findings on MOB-NDR/Lats Binding Specificity
| Research System | Specific Interaction Pairs | Functional Consequences | Structural Insights |
|---|---|---|---|
| S. cerevisiae (RAM network) | Cbk1 (Ndr) binds Mob2 | Controls cell separation and polarized growth | NTR forms V-shaped helical hairpin; Discrete residues determine specificity |
| S. cerevisiae (MEN network) | Dbf2/Dbf20 (Lats) bind Mob1 | Regulates cytokinesis and mitotic exit | Mob1-specific recognition motif identified |
| Human systems | NDR1/NDR2 bind MOB2 | Stimulates kinase activity; Roles in proliferation and morphogenesis | Association dramatically enhances catalytic activity |
| Human systems | LATS1/2 bind MOB1 | Regulates YAP/TAZ in Hippo signaling | Conserved interface architecture with specificity determinants |
Functional Implications of Selective MOB Binding
The strict specificity in MOB-NDR/Lats interactions enables discrete functional outcomes within interconnected signaling networks. In budding yeast, the RAM and MEN pathways control distinct cellular processes with no evidence of functional overlap between Cbk1-Mob2 and Dbf2/20-Mob1 complexes, despite simultaneous cytosolic presence of all components [1]. This separation of function underscores the biological importance of maintained binding specificity.
In metazoan systems, the conservation of specific MOB-NDR/Lats interactions facilitates specialized regulatory functions within Hippo signaling networks. The MOB1-LATS partnership primarily regulates cell proliferation and organ size through control of YAP/TAZ transcriptional coactivators, while MOB2-NDR interactions influence distinct processes including neuronal morphogenesis and cell polarity [1] [3]. This functional segregation suggests that therapeutic strategies targeting these interfaces would need to preserve native specificity to avoid disruptive cross-talk between signaling pathways.
Experimental Protocols
Co-Immunoprecipitation Protocol for MOB-NDR Interaction Validation
Day 1: Experiment Design and Cell Preparation
- Begin by designing the experiment to include appropriate controls: (1) positive control with known interactors, (2) negative control with non-interacting proteins, and (3) specific perturbations testing your hypothesis [43].
- Fuse the bait protein (e.g., NDR kinase) to GFP and prey proteins (e.g., MOB1/MOB2) to high-affinity short peptide tags such as HA, FLAG, or V5. Use different tags when expressing multiple prey proteins [43].
- Culture and expand an appropriate number of cells (e.g., 5×10^6 HEK293T cells per co-IP sample) to ensure sufficient material [43].
Day 2: Cell Lysis and Protein Extraction
- Prepare fresh cell lysis buffer (e.g., 50mM Tris-HCl pH 7.4, 150mM NaCl, 1mM EDTA, 1% NP-40) supplemented with protease and phosphatase inhibitors (1mM NaF, 1mM Na3VO4, 1mM sodium pyrophosphate) [43].
- Wash cells with ice-cold PBS and lyse in lysis buffer (typically 30 minutes on ice with occasional vortexing). For nuclear proteins, brief sonication may help disrupt the nuclear membrane [39].
- Clarify the lysate by centrifugation at 13,000 × g for 15 minutes at 4°C. Transfer the supernatant to a new tube and reserve 1-10% as "input" control [39] [43].
- Determine protein concentration and use approximately 300μg to 2mg total protein per IP, adjusting based on target protein abundance [39].
Day 3: Immunoprecipitation and Wash Steps
- Pre-clear the lysate by incubating with protein G agarose resin for 30-60 minutes at 4°C to reduce non-specific binding [43].
- Incubate the pre-cleared lysate with antibody against the bait protein (or GFP-tag) for 2-4 hours at 4°C with rotation. Antibody concentration must be optimized empirically.
- Add protein G agarose or magnetic beads and incubate for an additional 1-2 hours to capture immune complexes [40].
- Pellet beads magnetically or by brief centrifugation and wash 3-4 times with ice-cold lysis buffer with varying salt concentrations (150-500mM NaCl) to reduce non-specific binding [39] [40].
Day 4: Elution and Analysis
- Elute bound proteins using 2× SDS sample buffer (harsh elution) or gentle elution methods such as low-pH glycine buffer (0.1M, pH 2.5-3.0) or competitive elution with free peptide [40].
- Analyze eluates by SDS-PAGE and western blotting using antibodies against bait and prey proteins.
- For semi-quantitative analysis, perform densitometry measurements of band intensities and calculate the ratio of co-precipitated prey to bait protein, normalized to input controls [43].
Cross-linking Enhanced Co-IP for Transient Interactions
For capturing weak or transient MOB-NDR interactions, cross-linking enhanced Co-IP provides improved stabilization:
- Treat cells or freshly prepared lysates with membrane-permeable cross-linking agents such as dithiobis(succinimidyl propionate) (DSP) or bis(sulfosuccinimidyl) suberate (BS3) at concentrations ranging from 0.5-2mM for 30 minutes on ice [40].
- Quench the cross-linking reaction by adding Tris-HCl (pH 7.5) to a final concentration of 20-50mM and incubate for 15 minutes [40].
- Proceed with standard Co-IP protocol as described above, noting that cross-linked complexes may require longer washing times or higher salt concentrations to reduce background [40].
Pull-Down Assay for Direct Interaction Confirmation
- Express and purify recombinant NDR kinase NTR domain with an affinity tag (GST, His6, etc.) from E. coli or insect cells.
- Immobilize the tagged bait protein onto appropriate affinity resin (glutathione-sepharose for GST, Ni-NTA for His6) according to manufacturer recommendations.
- Incubate immobilized bait with purified MOB1 or MOB2 proteins (or cell lysates expressing these proteins) for 1-2 hours at 4°C with gentle mixing.
- Wash resin extensively with binding buffer (e.g., 50mM Tris-HCl pH 7.5, 150mM NaCl, 0.1% NP-40, 1mM DTT) to remove non-specifically bound proteins.
- Elute specifically bound proteins with tag-specific competitive agents (glutathione for GST, imidazole for His6) or SDS-PAGE sample buffer.
- Analyze eluates by SDS-PAGE and western blotting or mass spectrometry to identify interacting partners.
Visualization of Experimental Workflows and Signaling Pathways
Co-IP Experimental Workflow
MOB-NDR/Lats Specificity in Signaling
Research Reagent Solutions
Table 3: Essential Research Reagents for MOB-NDR Interaction Studies
| Reagent Category | Specific Examples | Function and Application |
|---|---|---|
| Cell Lines | HEK293T, Jurkat T-cells, HeLa | Protein expression and interaction studies; NDR1/2 localization [43] [3] |
| Expression Plasmids | pEGFP-N1, pcDNA3.1(+) | Mammalian expression of tagged proteins [43] |
| Affinity Tags | GFP, HA, FLAG, V5, GST, His6 | Protein detection and purification [39] [43] |
| Co-IP Beads | Protein A/G Agarose, Magnetic Dynabeads | Capture antibody-protein complexes [43] [42] |
| Lysis Buffers | NP-40, Triton X-100 based | Cell lysis while preserving interactions [43] [40] |
| Protease Inhibitors | Complete Protease Inhibitor Cocktail | Prevent protein degradation during processing [43] |
| Phosphatase Inhibitors | NaF, Na3VO4, β-glycerophosphate | Preserve phosphorylation states [43] |
| Cross-linkers | DSP, BS3 | Stabilize transient interactions [40] |
| Detection Antibodies | Anti-GFP, Anti-HA, Anti-NDR1/2, Anti-MOB1/2 | Western blot detection of bait and prey [43] [3] |
Technical Considerations and Troubleshooting
Optimization Strategies for MOB-NDR Studies
Successful application of Co-IP and pull-down assays to MOB-NDR interaction studies requires careful optimization of several technical parameters. Lysis buffer composition represents a critical variable, with non-ionic detergents like NP-40 or Triton X-100 at concentrations of 0.1-1% typically providing optimal balance between complete cell lysis and preservation of protein complexes [39] [40]. Buffer stringency should be adjusted through salt concentration (150-300mM NaCl) to minimize non-specific binding while maintaining physiological interactions.
Antibody selection significantly impacts Co-IP outcomes, with monoclonal antibodies generally providing higher specificity but potentially lower signal compared to polyclonal alternatives [41]. For tagged protein approaches, GFP nanobodies or commercial anti-GFP resins offer high affinity and consistency [43]. Appropriate controls are essential for interpreting results, particularly when studying specific binding preferences between MOB1 and MOB2 with their respective kinase partners. Essential controls include (1) isotype-matched non-specific antibody, (2) bait-only transfection, and (3) vector-only transfection to identify non-specific binding to beads or tags.
Addressing Technical Challenges
Common challenges in MOB-NDR interaction studies include weak or transient interactions, antibody cross-reactivity, and disruption of complexes during processing. Cross-linking approaches can stabilize transient interactions, with DSP and BS3 effectively preserving MOB-kinase associations without permanent denaturation [40]. For interactions that may be disrupted by standard lysis conditions, gentle detergents such as digitonin may better preserve complex integrity.
Non-specific binding presents particular challenges when studying homologous proteins like MOB1 and MOB2. Increasing wash stringency through elevated salt concentration (up to 500mM NaCl), adding non-ionic detergents (0.1% Tween-20), or including competitor proteins (BSA at 0.1-1mg/mL) can reduce background without disrupting specific interactions [40]. When working with low-abundance endogenous complexes, increasing starting material (up to 2mg total protein) and concentrating eluates before analysis can improve detection sensitivity.
Co-immunoprecipitation and pull-down assays remain foundational techniques for validating protein-protein interactions, with particular utility in delineating the specific binding relationships between MOB coactivators and NDR/Lats kinases. As structural insights reveal the molecular determinants of MOB1-MOB2 binding specificity, these biochemical techniques provide essential functional validation in cellular contexts. The continued refinement of these methods, including cross-linking approaches and quantitative analysis platforms, enhances their sensitivity and reliability for detecting subtle interaction differences.
Future methodological developments will likely focus on improving the capacity to study endogenous complexes without overexpression, capturing transient interactions with higher temporal resolution, and integrating these approaches with omics technologies for comprehensive interaction network analysis. As drug discovery efforts increasingly target protein-protein interfaces, particularly in Hippo signaling pathways, robust Co-IP and pull-down methodologies will remain essential for validating compound effects on specific MOB-kinase interactions. The technical guidelines presented here provide a framework for applying these powerful techniques to ongoing investigations of MOB-NDR/Lats binding specificity and its functional consequences in health and disease.
The Nuclear Dbf2-related (NDR) kinases, NDR1 and NDR2, are central components of evolutionarily conserved signaling pathways that control crucial cellular processes including cell cycle progression, the DNA damage response, and morphogenesis [4]. A defining characteristic of their regulation is their dependence on binding with Mps one binder (MOB) coactivator proteins [2]. This interaction is not monolithic; rather, the identity of the binding MOB protein dictates the functional outcome for the kinase. Specifically, MOB1 and MOB2 exhibit striking binding specificity and provoke divergent biological effects despite their structural similarities [4]. MOB1 binding to NDR kinases enhances their catalytic activity and promotes signaling through pathways such as the Hippo pathway [44]. In contrast, MOB2 competes with MOB1 for binding to the same N-terminal regulatory domain on NDR1/2, and the resulting MOB2/NDR complex is associated with diminished NDR kinase activity [4]. This competitive binding establishes a regulatory paradigm where the relative abundance and activation of MOB1 versus MOB2 can fine-tune NDR kinase signaling output. The central role of these kinases in cell proliferation and survival underscores the importance of accurately measuring their activity in a MOB-dependent context, a capability essential for both basic research and drug discovery efforts targeting this signaling axis.
The MOB-NDR Signaling Axis
Structural and Functional Dichotomy of MOB1 and MOB2
The functional divergence between MOB1 and MOB2 arises from their specific protein-protein interactions and the consequent effects on downstream signaling. MOB1A/B functions as a direct regulator of both NDR and LATS kinases within the Hippo signaling pathway, leading to kinase activation [4]. The activated LATS1/2 complex, in turn, phosphorylates and inhibits the YAP transcriptional coactivator, thereby suppressing cell proliferation and promoting apoptosis [23]. This places the MOB1/NDR/LATS complex as a critical tumor suppressor module.
MOB2, however, interacts specifically with NDR1/2 kinases but not with LATS1/2 [23]. Biochemically, MOB2 acts as a competitive inhibitor of MOB1-NDR binding. The formation of the MOB2/NDR complex is associated with a blockage of NDR kinase activation [4]. The biological consequences of this competition are significant. Research has shown that MOB2 knockout in hepatocellular carcinoma cells promotes cell migration and invasion, while its overexpression has the opposite effect [23]. Mechanistically, by competing with MOB1 for NDR binding, MOB2 appears to free up MOB1 to more effectively activate LATS1, leading to increased phosphorylation and inactivation of the oncoprotein YAP [23]. Therefore, MOB2's role extends beyond simple inhibition of NDR, positioning it as a positive regulator of the LATS/YAP limb of the Hippo pathway.
Table 1: Functional Comparison of MOB1 and MOB2 in NDR Kinase Signaling
| Feature | MOB1 | MOB2 |
|---|---|---|
| Primary Binding Partners | NDR1/2 and LATS1/2 kinases [4] | NDR1/2 kinases specifically [23] |
| Effect on NDR Kinase Activity | Stimulates activation [44] | Competes with MOB1, associated with diminished activity [4] |
| Role in Hippo Signaling | Core component; activates NDR and LATS kinases [4] | Indirect positive regulator of LATS/YAP; knockout decreases YAP phosphorylation [23] |
| Cellular Phenotypes | Cell cycle control, tumor suppression [4] | Regulates cell motility, DNA damage response, cell survival [4] [23] |
Visualizing the MOB-NDR Signaling Network
The competitive binding relationship between MOB1, MOB2, and NDR kinases, and their subsequent downstream effects, can be visualized in the following signaling pathway. This diagram integrates the key molecular interactions and functional outcomes, highlighting the distinct roles of MOB1 and MOB2.
Experimental Strategies for Measuring MOB-Dependent NDR Activation
Core Assay Principles and Platform Selection
Quantifying the activity of NDR kinases in response to MOB protein binding relies on measuring the kinase's ability to phosphorylate a substrate, typically detected by a change in signal (radioactivity, fluorescence, or luminescence). A critical consideration for any kinase assay is the concentration of ATP used, as it should reflect physiological conditions (∼1 mM) for the most biologically relevant assessment of inhibitor potency or activator efficacy [45]. The "HotSpot" assay is a prominent example of a radiometric filter-binding assay that meets this need and is well-suited for kinase profiling studies [46] [45].
Table 2: Key Kinase Activity Assay Platforms for MOB-NDR Research
| Assay Type | Detection Principle | Advantages | Disadvantages |
|---|---|---|---|
| Radiometric (HotSpot) | Measures transfer of 32P/33P from ATP to substrate using radioactive ATP [45]. | High sensitivity; universal application across kinases; uses physiologically relevant ATP [45]. | Requires handling and disposal of radioactive materials [45]. |
| Fluorescence-Based | Uses fluorophore-tagged substrates that emit light upon phosphorylation [45]. | Simple, cost-effective, suitable for high-throughput screening [45]. | Can experience compound interference and higher background; may use non-physiological substrates [45]. |
| Luminescence-Based | Measures ATP depletion via luciferase activity [45]. | Direct activity measurement; amenable to high-throughput [45]. | Indirect measurement; signal is coupled to ATP consumption rather than phosphorylation directly. |
| Ligand Binding | Measures compound-kinase binding affinity, not direct activity [45]. | Sensitive and can be high-throughput [46]. | Does not measure functional kinase activity; limited to kinases with known ligands [45]. |
Detailed Protocol: Co-Immunoprecipitation and Kinase Assay
This protocol outlines a standard method for assessing how MOB1 or MOB2 binding affects NDR kinase activity in a cellular context.
Objective: To isolate specific MOB-NDR complexes from cell lysates and measure the subsequent kinase activity of NDR.
Methodology:
Plasmid Transfection and Cell Lysis:
- Transfect HEK 293 or COS-7 cells with plasmids encoding tagged versions of NDR1 (or NDR2) and either MOB1 or MOB2. Common tags include HA (hemagglutinin) and myc [13].
- After 24-48 hours, lyse the cells in a non-denaturing lysis buffer (e.g., RIPA buffer) supplemented with protease and phosphatase inhibitors to preserve protein interactions and phosphorylation status.
Co-Immunoprecipitation (Co-IP):
- Incubate the clarified cell lysate with an antibody specific to the tag on your NDR kinase (e.g., anti-HA antibody) [13].
- Add Protein A/G beads to capture the antibody-kinase complex. Include extensive washing steps with lysis buffer to remove non-specifically bound proteins.
In Vitro Kinase Reaction:
- Resuspend the washed beads in a kinase reaction buffer containing MgCl₂, MnCl₂, and ATP [13]. The specific concentrations of detergents and MnCl₂ can significantly impact results and should be optimized [45].
- To measure kinase activity, add a suitable substrate. The reaction is often initiated by adding ATP and incubating at 30°C for 30-60 minutes.
- Stop the reaction by adding SDS-PAGE loading buffer.
Detection and Quantification:
- Resolve the proteins by SDS-PAGE and transfer to a membrane for immunoblotting.
- Use phospho-specific antibodies to detect phosphorylation of the NDR kinase itself (e.g., at Thr444 for NDR1) or of the added substrate [13].
- Quantify the band intensity to compare the kinase activity between MOB1- and MOB2-bound NDR complexes.
Detailed Protocol: Investigating MOB-NDR Specificity via Mutagenesis
To pinpoint the molecular determinants of MOB binding specificity, structure-guided mutagenesis is a powerful approach, informed by crystal structures of kinase-MOB complexes [2].
Objective: To identify critical residues in NDR kinases that dictate selective binding to MOB1 versus MOB2.
Methodology:
Site-Directed Mutagenesis:
Functional Analysis of Mutants:
- Co-transfect cells with wild-type or mutant NDR1 and either MOB1 or MOB2.
- Perform co-IP experiments as described in Section 3.2 to test if the mutations disrupt binding to one MOB protein while preserving binding to the other.
- Measure the kinase activity of the immunoprecipitated complexes. A successful specificity mutant would, for example, bind MOB2 but not MOB1, and its kinase activity would remain low, consistent with MOB2's inhibitory role.
The experimental workflow for these protocols is summarized below.
The Scientist's Toolkit: Essential Research Reagents
Successful investigation of MOB-dependent NDR activation requires a curated set of high-quality reagents. The table below lists essential tools for researchers in this field.
Table 3: Key Research Reagents for MOB-NDR Kinase Studies
| Reagent / Tool | Function and Application | Key Details |
|---|---|---|
| Active, Recombinant NDR Kinases | Core enzymatic component for in vitro activity and binding assays. | Human NDR1 and NDR2; quality should be verified by functional assay [46]. |
| Recombinant MOB1 & MOB2 Proteins | Essential coactivators for studying binding specificity and kinase stimulation in vitro. | Purified human proteins; used to reconstitute functional complexes with NDR kinases [44]. |
| Phospho-Specific Antibodies | Critical for detecting activation-loop phosphorylation of NDR kinases in cellular assays. | e.g., anti-NDR1 pThr444; validates kinase activation status in Western blotting [13]. |
| Tag-Specific Antibodies | Enable immunoprecipitation and detection of transfected proteins in cellular studies. | Anti-HA, anti-myc; used for Co-IP and Western blot analysis of tagged NDR and MOB proteins [13]. |
| Kinase Profiling Services (HotSpot) | Provides unbiased, functional screening of compound activity or kinase specificity across hundreds of kinases. | Uses radiometric assay at physiological ATP; ideal for comprehensive selectivity profiling [46] [45]. |
| Validated Kinase Inhibitors | Tool compounds for pathway validation and as controls in kinase activity assays. | e.g., compounds with known selectivity profiles from large-scale inhibitor screens [46]. |
The precise measurement of NDR kinase activity, with a specific focus on the dichotomous regulatory effects of MOB1 and MOB2, is a cornerstone for understanding a critical signaling node in cell biology and cancer research. The experimental strategies detailed herein—from robust activity assays and co-immunoprecipitation protocols to structure-informed mutagenesis—provide a framework for dissecting this complex. The competitive binding between MOB1 and MOB2 for the NDR kinase represents a finely-tuned regulatory mechanism controlling cell fate decisions. As research progresses, the application of these techniques, particularly those employing physiologically relevant conditions and comprehensive profiling, will be indispensable for the discovery and development of targeted therapies that aim to modulate this pivotal kinase pathway.
CRISPR/Cas9 and RNAi for Functional Genetic Studies
Functional genetic studies are fundamental to modern biological research, enabling scientists to decipher the roles of specific genes in cellular processes. Two powerful technologies, RNA interference (RNAi) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9, have revolutionized this field. While RNAi achieves gene silencing at the mRNA level (knockdown), CRISPR-Cas9 enables permanent disruption at the DNA level (knockout). The choice between these systems significantly impacts experimental outcomes, particularly in complex signaling pathways. Research into the binding specificity of MOB1 versus MOB2 coactivators for Nuclear Dbf2-related (NDR) kinases provides an excellent context for comparing these technologies. NDR kinases are crucial regulators of cell proliferation, morphogenesis, and Hippo signaling pathways, with MOB coactivators serving as essential binding partners that determine kinase activity and functional specificity [15] [1]. This technical guide examines the strategic application of RNAi and CRISPR-Cas9 for investigating such specific protein-protein interactions, providing detailed methodologies and comparative analysis for research scientists and drug development professionals.
RNA Interference (RNAi): mRNA-Level Knockdown
RNAi is an evolutionarily conserved biological mechanism that mediates gene silencing through sequence-specific mRNA degradation or translational repression. The process begins with the introduction of double-stranded RNA (dsRNA) into cells, which is recognized and cleaved by the endonuclease Dicer into small interfering RNAs (siRNAs) or microRNAs (miRNAs) approximately 21-25 nucleotides in length. These small RNAs are then loaded into the RNA-induced silencing complex (RISC), where the guide strand directs sequence-specific binding to complementary mRNA targets. Perfect complementarity leads to Argonaute-mediated mRNA cleavage, while imperfect matches result in translational repression [47].
The experimental workflow for RNAi involves:
- Design of highly specific siRNAs or miRNAs targeting the gene of interest
- Delivery into cells via plasmid vectors, synthetic siRNAs, PCR products, or in vitro transcribed RNAs
- Analysis of silencing efficiency through qRT-PCR (mRNA levels), immunoblotting (protein levels), or phenotypic assessment [47]
CRISPR-Cas9: DNA-Level Genome Editing
The CRISPR-Cas9 system functions as an adaptive immune mechanism in bacteria that has been repurposed for precise genome editing in eukaryotic cells. The technology requires two components: a guide RNA (gRNA) that provides targeting specificity through complementary base pairing, and the CRISPR-associated endonuclease (Cas9) that creates double-strand breaks in DNA. The most commonly used nuclease, SpCas9 from Streptococcus pyogenes, contains two functional lobes: a recognition lobe that verifies target complementarity and a nuclease lobe that cleaves DNA [47].
Upon creating a double-strand break, cells primarily utilize the error-prone non-homologous end joining (NHEJ) repair pathway, often resulting in insertions or deletions (indels) that disrupt the coding sequence and generate knockout alleles. Alternatively, researchers can harness the homology-directed repair (HDR) pathway with donor DNA templates to introduce specific sequence changes (knock-ins) [47].
The standard CRISPR-Cas9 workflow comprises:
- gRNA design using specialized bioinformatics tools to maximize on-target efficiency and minimize off-target effects
- Component delivery via plasmids, in vitro transcribed RNAs, or preassembled ribonucleoprotein (RNP) complexes
- Validation of editing efficiency through methods like T7 endonuclease assay, tracking of indels by decomposition (TIDE), or next-generation sequencing [48] [47]
Application in MOB-NDR Kinase Specificity Research
Biological Context: MOB-NDR Interactions
NDR kinases form a conserved subgroup of AGC kinases that function as essential regulators of growth, morphogenesis, and proliferation in eukaryotes. These kinases require binding with MOB (Mps one binder) coactivator proteins for full activation and proper subcellular localization. In humans, the NDR kinase family includes NDR1/STK38 and NDR2/STK38L, which preferentially bind MOB2 proteins, and LATS1/2 kinases, which associate with MOB1 proteins [15] [1].
Structural studies have revealed that NDR/Lats kinases contain a characteristic N-terminal regulatory (NTR) region that mediates specific interaction with MOB cofactors. The interface between NDR-NTR and MOB forms a structural platform that organizes the kinase hydrophobic motif and facilitates allosteric regulation [1]. This specific binding is crucial for proper kinase function, as evidenced by research showing that MOB1-DBF2 and MOB2-COT1 form distinct functional complexes in Neurospora crassa with separate roles in septation and tip growth, respectively [49].
Table 1: MOB Family Proteins and Their NDR Kinase Partners
| MOB Class | Representative Members | Preferred Kinase Partners | Cellular Functions |
|---|---|---|---|
| Class I | MOB1A, MOB1B | LATS1/2, DBF2/20 | Hippo signaling, mitotic exit, cytokinesis [15] |
| Class II | MOB2A, MOB2B | NDR1/2, COT1, CBK1 | Cell morphogenesis, polarity, neuronal development [15] [49] |
| Class III | MOB3 | Not well characterized | Poorly understood, may have kinase-independent functions [15] |
| Class IV | MOB4/Phocein | STRIPAK complex (phosphatase) | Antagonizes Hippo signaling [15] |
Experimental Strategies for Binding Specificity Studies
Investigating MOB-NDR binding specificity requires complementary approaches using both RNAi and CRISPR-Cas9 technologies:
RNAi-Based Approaches:
- Knockdown studies to assess functional compensation between MOB paralogs
- Transient suppression to study essential genes where complete knockout is lethal
- Dose-response relationships by titrating siRNA concentrations to achieve partial knockdown
- High-throughput screening with arrayed siRNA libraries targeting MOB and NDR family members
CRISPR-Cas9 Approaches:
- Constitutive knockout of specific MOB or NDR genes to establish null phenotypes
- Domain-specific mutagenesis to disrupt binding interfaces while preserving kinase activity
- Endogenous tagging for localization and interaction studies
- CRISPRi for reversible suppression without permanent genetic alteration
A notable example of RNAi application in Hippo pathway research comes from a genome-wide screen that identified COPI complex subunits as regulators of YAP localization in LATS1/2 double-knockout cells, revealing Hippo-independent mechanisms of YAP regulation [50].
Comparative Analysis: RNAi vs. CRISPR-Cas9
Technical Performance Parameters
Table 2: Quantitative Comparison of RNAi and CRISPR-Cas9 Technologies
| Parameter | RNAi | CRISPR-Cas9 |
|---|---|---|
| Mechanism of action | mRNA degradation/translational repression [47] | DNA cleavage with subsequent indel formation [47] |
| Level of intervention | Transcriptional/translational (knockdown) [47] | Genomic (knockout) [47] |
| Typical efficiency | 70-90% mRNA reduction (highly variable) [47] | 50-95% editing efficiency (more consistent) [47] |
| Reversibility | Transient/reversible | Permanent (except CRISPRi) |
| Off-target effects | Significant (sequence-dependent and independent) [47] | Moderate (improved with high-fidelity Cas variants) [51] [47] |
| Duration of effect | Days to weeks | Permanent (constitutive editing) |
| Multiplexing capacity | Moderate (limited by delivery efficiency) | High (multiple gRNAs) |
| Screening applications | Arrayed and pooled formats | Primarily pooled, arrayed gaining traction [48] |
Strategic Considerations for MOB-NDR Research
The investigation of MOB-NDR kinase interactions presents specific challenges that influence technology selection:
Advantages of RNAi:
- Enables study of essential genes where complete knockout is lethal—particularly relevant for core Hippo pathway components [47]
- Facilitates assessment of dose-dependent effects by achieving partial reduction of gene expression
- Allows reversible suppression for studying temporal requirements of MOB-NDR interactions
- Potentially better for studying paralog compensation within MOB families
Advantages of CRISPR-Cas9:
- Complete ablation of specific MOB or NDR isoforms eliminates concerns about residual protein function
- More reliable for interpreting phenotypic consequences due to more complete gene disruption
- Enables precise editing of MOB binding interfaces in NDR kinases to map interaction domains
- Lower off-target rates provide higher confidence in phenotype-genotype correlations [47]
Recent advances in CRISPR technology, including the development of base editing and prime editing, offer additional precision for introducing specific point mutations to dissect MOB-NDR binding interfaces without creating double-strand breaks.
Experimental Protocols
RNAi Protocol for MOB/NDR Knockdown
Reagents Required:
- Validated siRNA oligonucleotides targeting MOB1, MOB2, NDR1, or NDR2
- Non-targeting control siRNA
- Appropriate transfection reagent
- Cell culture media and supplements
- Antibodies for validation (e.g., anti-MOB1, anti-NDR1/2)
Step-by-Step Methodology:
- Design and Selection: Select 3-5 different siRNA sequences per target gene focusing on conserved regions of MOB1 (e.g., NM001330302.1) or MOB2 (e.g., NM001142463.2). Avoid seed region matches to off-target transcripts.
- Cell Preparation: Plate appropriate cell lines (HEK293T, MCF7, or specialized Hippo pathway reporter cells) at 30-50% confluency in antibiotic-free medium.
- Transfection Complex Formation: Dilute siRNA to working concentration (typically 10-50 nM) in serum-free medium. Mix with transfection reagent at optimized ratios. Incubate 15-20 minutes at room temperature.
- Transfection: Add complexes to cells dropwise with gentle swirling. Include non-targeting siRNA and untreated controls.
- Incubation and Analysis: Harvest cells 48-96 hours post-transfection for:
- mRNA quantification by qRT-PCR using primers for MOB/NDR genes
- Protein analysis by immunoblotting
- Functional assays (kinase activity, co-immunoprecipitation, localization studies)
Validation Measures:
- Confirm knockdown efficiency at both mRNA (70-90% reduction expected) and protein levels
- Assess specificity by monitoring related paralogs (e.g., ensure MOB1 knockdown doesn't affect MOB2)
- Include rescue experiments with siRNA-resistant cDNA constructs where possible
CRISPR-Cas9 Protocol for MOB/NDR Knockout
Reagents Required:
- Validated gRNA sequences cloned into appropriate expression vector
- Cas9 expression plasmid or recombinant protein
- Selection antibiotics (puromycin, blasticidin) or fluorescence markers
- PCR reagents for genotyping
- T7 endonuclease I or similar detection reagent
Step-by-Step Methodology:
- gRNA Design and Cloning: Design 3-5 gRNAs targeting early exons of MOB1 (e.g., Chr9: 27426370-27426389) or MOB2 (e.g., Chr11: 72738291-72738310). Clone into Cas9-compatible vectors (e.g., lentiCRISPRv2).
- Cell Transduction/Transfection:
- For lentiviral delivery: Produce lentivirus in packaging cells, transduce target cells at MOI 0.4-0.6 [48]
- For RNP delivery: Complex synthetic gRNA with recombinant Cas9 protein, deliver via electroporation or lipid nanoparticles
- Selection and Clonal Isolation: Apply appropriate selection 24-48 hours post-transduction for 5-7 days. For clonal isolation, single-cell sort into 96-well plates or perform limiting dilution.
- Screening and Validation:
- Extract genomic DNA from potential clones
- PCR-amplify target regions and analyze by T7E1 assay or sequencing
- Confirm knockout by immunoblotting and functional assays
- Sequence verified clones to characterize specific indel mutations
Validation Measures:
- Confirm complete absence of target protein by immunoblotting
- Sequence target loci to verify frameshift mutations
- Perform functional validation (e.g., loss of MOB1 should disrupt LATS activation)
- Check for compensatory upregulation of related family members
Research Reagent Solutions
Table 3: Essential Research Reagents for MOB-NDR Studies
| Reagent Category | Specific Examples | Applications | Technical Notes |
|---|---|---|---|
| RNAi Reagents | MISSION shRNA libraries, Silencer Select siRNAs [48] | Targeted knockdown, high-throughput screening | Include multiple constructs per target; validate extensively |
| CRISPR Tools | GeCKO libraries, Synthetic sgRNAs, Cas9-expressing cell lines [48] | Knockout generation, functional genomics | RNP format increases efficiency; use validated Cas9 cell lines |
| Cell Lines | RPE1-hTERT, HEK293T, MCF7, Hippo pathway reporter lines [50] [48] | Functional assays, signaling studies | Select lines with functional Hippo/NDR pathways; consider LATS DKO for Hippo-independent studies [50] |
| Validation Antibodies | Anti-MOB1, Anti-NDR1/2, Anti-YAP/TAZ, Phospho-specific antibodies [50] [52] | Western blot, immunofluorescence, IP | Verify specificity with knockout controls; phospho-antibodies for activity assessment |
| Specialized Assays | TEAD reporter assays, Kinase activity assays, Co-IP protocols [50] [53] | Functional validation, interaction studies | Include appropriate controls; YAP localization as functional readout [53] |
Signaling Pathway Visualization
Figure 1: MOB-NDR Kinase Signaling Networks. This diagram illustrates the interconnected signaling pathways involving MOB coactivators and NDR kinases, showing how MOB1 and MOB2 form distinct complexes with LATS and NDR kinases, respectively, to regulate YAP/TAZ activity and cellular processes [15] [53].
The strategic selection between RNAi and CRISPR-Cas9 technologies for investigating MOB-NDR kinase specificity depends on multiple experimental factors, including the biological question, required perturbation level, and technical constraints. RNAi remains valuable for studying essential genes, establishing dose-response relationships, and conducting reversible suppression experiments. In contrast, CRISPR-Cas9 offers more complete and specific gene disruption, making it preferable for determining null phenotypes and establishing unambiguous genotype-phenotype relationships.
Future methodological developments will likely enhance both technologies. Advances in CRISPR inhibition (CRISPRi) enable reversible transcriptional repression without DNA cleavage, potentially combining the benefits of both approaches. Similarly, improved RNAi design algorithms and chemical modifications continue to reduce off-target effects. For researchers studying MOB-NDR kinase interactions, a combined approach utilizing CRISPR-generated knockout cell lines complemented with RNAi-based acute suppression and rescue experiments provides the most rigorous experimental framework. This multi-faceted methodology ensures that observed phenotypes result from specific perturbation of the targeted interaction rather than compensatory mechanisms or off-target effects, ultimately leading to more reliable conclusions about MOB-NDR binding specificity and its functional consequences in cellular signaling.
Cellular localization and membrane recruitment are central to kinase signaling regulation. For the NDR kinase family (NDR1/2), membrane translocation activates critical pathways controlling cell cycle progression, DNA damage response, and polarity [4] [13]. This process is governed by MOB proteins (MOB1 and MOB2), which exhibit distinct binding specificities and functional outcomes. MOB1 activates NDR kinases by promoting autophosphorylation, while MOB2 competes for NDR binding and may suppress kinase activity [4] [13]. This technical guide details methodologies and mechanisms for studying MOB-driven membrane recruitment of NDR kinases, providing a framework for targeted drug discovery.
Molecular Mechanisms of MOB-NDR Signaling
2.1 MOB1 vs. MOB2: Binding Specificity and Functional Divergence MOB proteins allosterically regulate NDR kinases, but their structural differences dictate signaling outcomes:
- MOB1 forms complexes with both NDR and LATS kinases, enhancing NDR catalytic activity through phosphorylation at Ser281/Thr444 (NDR1) or Ser282/Thr442 (NDR2) [13].
- MOB2 binds specifically to NDR kinases (not LATS) and may inhibit activation by competing with MOB1 [4]. Biochemical studies show MOB2/NDR complexes correlate with reduced NDR activity, suggesting a dominant-negative role [4].
2.2 Membrane Recruitment as an Activation Trigger NDR kinases are predominantly cytoplasmic, and their activation requires membrane translocation. Key steps include:
- MOB Localization: MOB proteins recruit NDR to the plasma membrane.
- Phosphorylation: Membrane-bound NDR undergoes phosphorylation by upstream kinases (e.g., MST1/2) and autophosphorylation.
- Effector Engagement: Activated NDR phosphorylates substrates like p21/Cip1 and c-Myc to regulate G1/S cell cycle progression [4].
Table 1: Quantitative Comparison of MOB1 vs. MOB2 Functions
| Feature | MOB1 | MOB2 |
|---|---|---|
| NDR Binding | Yes (activates) | Yes (inhibits) |
| LATS Binding | Yes | No |
| Kinase Activity | Increases NDR phosphorylation | Suppresses NDR activation |
| Cellular Localization | Plasma membrane/cytoplasm | Plasma membrane/cytoplasm |
| Role in DDR | Indirect | Direct (via RAD50/MRN complex) |
2.3 Pathway Crosstalk and Disease Relevance
- DNA Damage Response (DDR): MOB2 stabilizes the MRN complex (MRE11-RAD50-NBS1) to facilitate ATM kinase recruitment and G1/S checkpoint activation [4].
- Cell Polarity: In S. pombe, the NDR homolog Orb6 phosphorylates Cdc42 GAP Rga3 to regulate exploratory Cdc42 dynamics during stress [54].
- Cancer Pathways: NDR2 overexpression promotes tumor progression in lung cancer by modulating vesicle trafficking and autophagy [8].
Experimental Protocols for Membrane Recruitment Studies
3.1 Live-Cell Imaging of Protein Translocation Objective: Visualize real-time recruitment of NDR kinases to membranes.
Methodology:
- Construct Design:
- Fuse NDR1/2 and MOB proteins with fluorescent tags (e.g., GFP, mCherry).
- Generate membrane-targeted MOBs using N-terminal myristoylation/palmitylation motifs (e.g., mp-HA from Lck tyrosine kinase) [13].
- Transfection: Use COS-7, HEK293, or U2-OS cells transfected with Lipofectamine 2000.
- Stimuli: Treat cells with:
- Okadaic acid (1 μM, 60 min) to inhibit PP2A and enhance phosphorylation.
- Phorbol esters (e.g., TPA, 100 ng/mL) to activate PKC-driven membrane translocation.
- Imaging: Track fluorescence redistribution via confocal microscopy pre-/post-stimulation.
Key Controls:
- Kinase-dead NDR mutants (K118A-NDR1).
- Phospho-deficient mutants (T444A-NDR1).
3.2 Co-Immunoprecipitation (Co-IP) for Complex Formation Objective: Validate MOB-NDR interactions and phosphorylation status.
Methodology:
- Lysis: Use RIPA buffer supplemented with phosphatase/protease inhibitors.
- Antibodies:
- Anti-NDR1/2 (CT antibody or phospho-specific anti-T444-P).
- Anti-MOB1A/B or MOB2 (commercial monoclonals).
- Pull-Down: Incubate lysates with antibody-bound beads (4°C, 4 h).
- Immunoblotting: Probe for:
- Total NDR (anti-NDR CT).
- Phospho-NDR (anti-Ser281-P/Thr444-P).
- MOB proteins (anti-HA/Myc).
Quantification: Measure band intensity to calculate phosphorylation stoichiometry.
3.3 Inducible Membrane Recruitment Systems Objective: Control MOB localization temporally using chemically induced dimerization.
Methodology:
- Design: Fuse hMOB1A to FRB domain and membrane anchor (e.g., C1 domain of PKCα).
- Transfection: Co-express with NDR-GFP in HEK293 cells.
- Induction: Add rapamycin (100 nM) to recruit hMOB1A to the membrane.
- Kinase Assay: Monitor NDR phosphorylation via immunoblotting at 0, 5, 15, and 30 min.
The Scientist’s Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Membrane Recruitment Studies
| Reagent | Function | Example/Catalog |
|---|---|---|
| MP-Tagged MOBs | Directs MOB proteins to the plasma membrane for recruitment assays | pcDNA3-mp-HA-hMOB1A [13] |
| Phospho-Specific Antibodies | Detects active NDR kinases (pT444/pS281) | Anti-NDR1 pT444 (custom) [13] |
| Okadaic Acid | PP2A inhibitor that enhances NDR phosphorylation | Sigma-Aldrich O9381 |
| CRBN-DDB1ΔB Construct | E3 ligase component for molecular glue studies (e.g., pomalidomide screening) | Recombinant FLAG-tagged protein [55] |
| SpyTag003-SpyCatcher003 | Covalent labeling for BLI-based binding assays | PMC12247212 [56] |
Signaling Pathway and Workflow Visualizations
MOB-NDR Membrane Recruitment and Activation Pathway
Title: MOB-NDR Membrane Activation Pathway
Experimental Workflow for Membrane Recruitment Assay
Title: Membrane Recruitment Experimental Workflow
Discussion and Future Directions
The mechanistic insights into MOB-NDR signaling have translational implications:
- Cancer Therapeutics: NDR2 inhibition may suppress lung adenocarcinoma metastasis by disrupting vesicle trafficking [8].
- Neurological Disorders: Aberrant NDR signaling contributes to tau hyperphosphorylation in Alzheimer's disease [57].
- Drug Screening: SpyBLI pipelines enable high-throughput kinetic measurements of MOB-NDR interactions using cell-free expression systems [56].
Future work should address:
- Structural Basis of Specificity: Resolve MOB1-NDR vs. MOB2-NDR co-crystal structures.
- Single-Cell Dynamics: Develop biosensors for real-time MOB-NDR activity tracking.
- Therapeutic Targeting: Design molecular glues that modulate MOB-NDR complex formation [55].
This guide integrates technical protocols with mechanistic insights to advance research on MOB-NDR signaling in disease contexts.
Phospho-Specific Antibodies for Monitoring Activation Status
Phospho-specific antibodies (PSSAs) represent a cornerstone technology in molecular cell biology, enabling researchers to investigate the dynamic regulation of protein phosphorylation in situ. These specialized reagents are uniquely designed to distinguish between phosphorylated and non-phosphorylated states of proteins at specific residues, providing unprecedented insight into signaling pathway activation and protein function regulation within cellular contexts. The development of PSSAs has transformed our understanding of intracellular signaling, moving beyond biochemical studies of homogenized tissues to allowing precise examination of phosphorylation events within the spatially complex structures of cells and tissues [58].
In the specific context of NDR kinase research, phospho-specific antibodies have proven indispensable for elucidating the complex activation mechanisms of these crucial regulatory enzymes. The NDR kinase family, including human NDR1 and NDR2, requires phosphorylation at conserved sites for full activation and proper cellular function. These kinases represent a subfamily of AGC protein kinases with established roles in cell cycle progression, cell morphology, and * Hippo pathway signaling*, with particular relevance to cancer biology when dysregulated [13] [59]. This technical guide will explore the application of phospho-specific antibodies for monitoring NDR kinase activation, with specific emphasis on distinguishing between MOB1 and MOB2 coactivator binding specificity, a critical determinant of NDR kinase function and downstream signaling outcomes.
NDR Kinase Activation and MOB Protein Specificity
NDR Kinase Activation Mechanism
NDR kinases undergo a multistep activation process requiring phosphorylation at two critical residues and association with MOB coactivator proteins. Research has established that human NDR1 requires phosphorylation at Ser281 (located within the activation loop) and Thr444 (located within the hydrophobic motif at the C-terminal regulatory domain) for full enzymatic activity [13] [26]. The analogous sites in NDR2 are Ser282 and Thr442. Phosphorylation at these sites occurs through distinct mechanisms: Ser281/282 undergoes autophosphorylation in a Ca2+-dependent manner, while Thr444/442 is phosphorylated by upstream kinases, particularly Ste20-like kinases such as MST3 [26].
The activation mechanism involves relief from autoinhibition mediated by an atypically long activation segment that blocks substrate binding and stabilizes an inactive kinase conformation in the non-phosphorylated state [24]. Recent structural studies have revealed that this autoinhibitory segment stabilizes a non-productive position of helix αC, with mutations within this region dramatically enhancing in vitro kinase activity [24]. The complete activation process therefore involves coordinated phosphorylation events, MOB coactivator binding, and conformational changes that release autoinhibition.
MOB Binding Specificity in NDR Kinase Regulation
MOB proteins function as essential coactivators and regulatory subunits for NDR/LATS family kinases, with distinct binding specificities that determine functional outcomes. Structural and biochemical studies have established that NDR kinases specifically associate with MOB2 proteins, while LATS kinases preferentially bind MOB1 proteins [1] [59]. This specific pairing is maintained across evolutionary boundaries from yeast to humans, indicating its fundamental importance to kinase regulation and function.
The molecular basis for this binding specificity resides in the N-terminal regulatory (NTR) region of NDR/LATS kinases, which forms a conserved structural platform that mediates selective interaction with distinct MOB family members [1]. Structural analyses of Saccharomyces cerevisiae Cbk1NTR-Mob2 and Dbf2NTR-Mob1 complexes have revealed that specificity is determined by discrete sites rather than being broadly distributed across the interaction interface [1]. The MOB-organized NTR appears to mediate association of the hydrophobic motif with an allosteric site on the N-terminal kinase lobe, providing a distinctive kinase regulation mechanism [1].
Functionally, MOB1 and MOB2 binding produces divergent regulatory outcomes for NDR kinases. While MOB1A binding enhances NDR kinase activity, leading to fully active enzymes, MOB2 binding competes with MOB1 and is associated with diminished NDR activity [4]. This competitive binding relationship suggests a model where the relative abundance and activation of different MOB proteins can fine-tune NDR kinase signaling output in response to cellular conditions.
Table 1: Key Phosphorylation Sites in Human NDR Kinases
| Kinase | Activation Loop Site | Hydrophobic Motif Site | Required for Activation | Phosphorylation Mechanism |
|---|---|---|---|---|
| NDR1 | Ser281 | Thr444 | Yes | Ser281: Autophosphorylation [13]Thr444: Upstream kinase (e.g., MST3) [26] |
| NDR2 | Ser282 | Thr442 | Yes | Ser282: Autophosphorylation [13]Thr442: Upstream kinase (e.g., MST3) [26] |
Phospho-Specific Antibodies in NDR Kinase Research
Development and Validation of Phospho-Specific Antibodies
The development of phospho-specific antibodies typically involves immunization strategies using synthetic phosphorylated peptides corresponding to the target phosphorylation site. Early approaches relied on enzymatic phosphorylation of peptide antigens, but advances in peptide chemistry now enable direct chemical phosphorylation, significantly improving efficiency and yield [58]. For polyclonal antibody production, antisera are typically subjected to a two-step purification process: first removing antibodies that bind to the dephosphorylated antigen using a dephospho-peptide affinity column, followed by positive selection of the flow-through on a phospho-peptide column [58].
Validation of phospho-specific antibodies requires rigorous testing to establish specificity and reliability. For phospho-specific flow cytometry antibodies, ThermoFisher Scientific's Invitrogen eBioscience recommends a comprehensive validation approach including [60]:
- Pathway-specific tests: Verifying that staining occurs only in cells where the specific pathway of interest has been activated
- Cell-type specific tests: Confirming that phosphorylation-specific staining is observed only in cell types where the target protein is expressed
- Western blot correlation: Whenever possible, confirming the presence of bands of appropriate size in stimulated/treated cells but not in unstimulated controls
- Inhibitor studies: Using pathway inhibitors to verify expected increases or decreases in signal intensity
- Buffer compatibility tests: Evaluating antibody performance across different fixation and permeabilization buffer systems
For NDR kinase research, specific phospho-antibodies have been developed against the critical phosphorylation sites. For instance, anti-NDR1 phospho-Thr444 antibodies were validated using peptide competition assays, with specificity confirmed by blocked staining in the presence of the phospho-peptide (KDWVFINYTpYKRFEG) but not the dephospho-peptide (KDWVFINYTYKRFEG) [13].
Application to MOB1 vs MOB2 Binding Studies
Phospho-specific antibodies enable researchers to investigate how MOB coactivator binding influences NDR kinase phosphorylation status and activation. Studies utilizing these reagents have demonstrated that membrane targeting of either NDR or MOB proteins results in constitutive kinase activation through phosphorylation at both Ser281 and Thr444 sites [13]. Strikingly, using an inducible membrane translocation system for MOB1A, researchers demonstrated that NDR phosphorylation and activation at the membrane occurs within minutes after association of MOB1 with membranous structures [13].
The differential effects of MOB1 versus MOB2 binding on NDR kinase phosphorylation can be monitored using phospho-specific antibodies. While MOB1 binding promotes NDR activation and phosphorylation, MOB2 appears to compete with MOB1 for NDR binding and is associated with reduced NDR kinase activity [4]. This competitive relationship creates a balance mechanism where the relative expression and activation of MOB1 versus MOB2 can fine-tune NDR signaling output.
Table 2: Phospho-Specific Antibodies for NDR Kinase Research
| Target Phospho-Site | Antibody Name/Clone | Application Techniques | Key Validation Data | Utility in MOB Studies |
|---|---|---|---|---|
| NDR1 p-Thr444 | Custom anti-T444-P [13] | Western blot, Immunofluorescence | Specificity confirmed by peptide competition [13] | Monitoring HM phosphorylation dependent on MOB binding |
| NDR1 p-Ser281 | Custom anti-S281-P [13] | Western blot, Immunofluorescence | Correlation with kinase activity assays [13] | Detecting autophosphorylation enhanced by MOB1 |
| NDR2 p-Thr442 | Custom anti-T442-P [26] | Western blot | Abolished by MST3 knockdown [26] | Assessing upstream kinase input to MOB/NDR complex |
| NDR2 p-Ser282 | Custom anti-S282-P [26] | Western blot | Correlation with OA-induced activation [26] | Measuring autophosphorylation in MOB complexes |
Experimental Protocols for Monitoring NDR Activation
Cell Culture and Treatment Protocols
Cell lines commonly used in NDR kinase research include COS-7, U2-OS, HEK 293, and HeLa cells, typically maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum [13]. For phosphorylation studies, cells are typically plated at consistent confluence (e.g., 3 × 10^5 cells/6-cm dish) and transfected the following day using standard transfection reagents such as Fugene 6 (Roche) or Lipofectamine 2000 (Invitrogen) [13].
To induce NDR kinase activation, researchers employ several treatment approaches:
- Okadaic acid (OA) treatment: Cells are treated with 1 μM OA for 60 minutes to inhibit protein phosphatase 2A (PP2A), resulting in enhanced NDR phosphorylation and activation [13]
- Serum starvation and stimulation: Cells may be serum starved for 2 hours prior to transfection, followed by overnight serum starvation before stimulation with 100 ng/ml 12-O-tetradecanoylphorbol 13-acetate (TPA) [13]
- MST3 co-expression: To specifically enhance hydrophobic motif phosphorylation, researchers co-express wild-type MST3 but not kinase-dead MST3KR [26]
Immunodetection and Analysis Methods
For western blot analysis of NDR phosphorylation, cell lysates are prepared using IP buffer containing protease and phosphatase inhibitors (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1 mM Na3VO4, 20 mM β-glycerol phosphate, 1 μM microcystin, 50 mM NaF, 0.5 mM PMSF, 4 μM leupeptin, and 1 mM benzamidine) [26]. Samples are resolved by 8-12% SDS-PAGE, transferred to polyvinylidene difluoride membranes, and probed with phospho-specific antibodies overnight at 4°C [13] [26].
For intracellular flow cytometry analysis of phosphorylation, cells are typically fixed with formaldehyde to stabilize the cell membrane, followed by permeabilization with detergent or alcohol to allow antibody access to intracellular epitopes [60]. Several buffer systems should be compared for optimal results, including IC Fixation and Permeabilization, Foxp3/Transcription Factor Buffer, and IC Fixation Buffer/Methanol [60]. Methanol permeabilization can destroy some epitopes, so surface staining should be performed with methanol-resistant fluorochromes when using methanol-based protocols.
Immunofluorescence and immunohistochemistry applications enable spatial resolution of NDR phosphorylation within cellular compartments. For immunohistochemistry on formalin-fixed paraffin-embedded (FFPE) tissue, standard antigen retrieval methods are employed followed by incubation with phospho-specific antibodies and appropriate detection systems [58].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for NDR Kinase Phosphorylation Studies
| Reagent Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Phospho-Specific Antibodies | Anti-NDR1 p-Thr444, Anti-NDR1 p-Ser281 [13] | Detecting specific phosphorylation events | Validate specificity via peptide competition and pathway modulation |
| Cell Lines | COS-7, HEK293, HeLa [13] | Cellular context for signaling studies | Select based on transfection efficiency and endogenous protein expression |
| Kinase Modulators | Okadaic acid (1μM) [13], MST3 expression constructs [26] | Manipulating phosphorylation status | Use appropriate controls (e.g., kinase-dead mutants) |
| Transfection Reagents | Fugene 6, Lipofectamine 2000 [13] | Introducing expression constructs | Optimize for cell type and viability |
| Lysis & Buffers | IP buffer with phosphatase inhibitors [26], Fixation/Permeabilization kits [60] | Protein extraction and cell preparation | Maintain phosphorylation status during processing |
| MOB Expression Constructs | MOB1A, MOB1B, MOB2 [13] [4] | Studying coactivator specificity | Consider tagged versions for detection and localization |
Signaling Pathway Visualization
NDR Kinase Activation Pathway
Diagram 1: NDR kinase activation pathway and MOB protein specificity. MST3 phosphorylates the hydrophobic motif (Thr444/442), enabling MOB1 binding and subsequent kinase activation. MOB2 competes with MOB1, modulating activation.
Experimental Workflow for Phosphorylation Monitoring
Diagram 2: Experimental workflow for monitoring NDR phosphorylation. Key reagents (red) and detection tools (blue) are shown with dashed lines at their points of application.
Phospho-specific antibodies provide powerful tools for deciphering the complex regulation of NDR kinases by MOB coactivators. Through rigorous validation and appropriate application across multiple detection platforms, these reagents enable precise monitoring of kinase activation status and facilitate understanding of how MOB1 versus MOB2 binding specificity directs NDR signaling outcomes. As research continues to elucidate the roles of NDR kinases in cell cycle regulation, DNA damage response, and autophagy, phospho-specific antibodies will remain essential tools for connecting kinase activation states to functional consequences in both physiological and pathological contexts.
Research Challenges and Strategic Approaches for MOB-NDR Studies
Overcoming Non-Specific Binding in Interaction Studies
The study of specific interactions between Mps one binder (MOB) proteins and Nuclear Dbf2-related (NDR) kinases presents a significant challenge in molecular biology due to the pervasive issue of non-specific binding (NSB). NSB occurs when proteins or analytes adhere to surfaces or interaction partners through non-biological mechanisms, potentially compromising data accuracy and leading to erroneous conclusions [61] [62]. Within the MOB protein family, MOB1 and MOB2 exhibit strikingly different binding specificities despite their structural similarities. MOB1 interacts with both NDR and LATS kinases, while MOB2 demonstrates exclusive binding specificity for NDR1/2 kinases without forming complexes with LATS kinases [4]. This precise interaction profile makes the MOB1 versus MOB2 paradigm particularly valuable for studying specific protein-protein interactions, yet also necessitates sophisticated methods to distinguish true biological interactions from non-specific background binding.
The biological significance of these specific interactions is substantial. MOB proteins function as crucial signal transducers in essential intracellular pathways through their regulatory interactions with serine/threonine protein kinases of the NDR/LATS family [4]. Research indicates that MOB2 competes with MOB1 for NDR binding, with the MOB1/NDR complex corresponding to increased NDR kinase activity, while the MOB2/NDR complex is associated with diminished NDR activity [4]. This competitive balance has profound implications for cellular processes, including cell cycle progression, DNA damage response (DDR), and Hippo signaling pathway regulation [4]. Overcoming NSB is therefore not merely a technical concern but a fundamental prerequisite for accurately understanding these critical biological mechanisms.
Understanding Non-Specific Binding in Protein Interaction Studies
Non-specific binding arises from multiple sources in experimental systems, each requiring distinct mitigation strategies. The material composition of solid surfaces represents a primary contributor, as different materials contain various functional groups that promote distinct interactions with analytes [62]. For instance, glassware surfaces abundant in silanol groups readily acquire negative charges, facilitating binding with positively charged molecules through ionic interactions. Conversely, polypropylene and polystyrene materials contain hydrophobic groups that preferentially bind to hydrophobic molecules [62].
Solution composition significantly influences NSB through multiple mechanisms. pH levels affect the dissociation state of compounds, determining whether analytes exist in dissociated or molecular forms, which subsequently impacts their interaction with solid surfaces [62]. Buffer salts can occupy binding sites on solid surfaces, while proteins and lipids in solution may bind with analytes, potentially weakening or eliminating analyte binding to solid surfaces [62]. Compound properties further contribute to NSB challenges, with hydrophobic, hydrophilic, and amphiphilic compounds each exhibiting distinct binding preferences through different mechanisms [62].
In the context of MOB-NDR kinase research, NSB presents particular challenges when working with complex biological samples. Surface plasmon resonance (SPR) studies have demonstrated that serum components produce heterogeneous and non-controllable NSB, which can obscure specific interaction signals and complicate data interpretation [61]. This problem is especially relevant when investigating MOB2's recently identified role in DNA damage response, where it interacts with RAD50 component of the MRN DNA damage sensor complex [4]. Accurate characterization of these interactions requires meticulous elimination of NSB to distinguish biologically relevant binding from experimental artifact.
Mathematical Approaches to Distinguish Specific from Non-Specific Binding
Advanced mathematical models provide powerful tools for deconvoluting specific binding from NSB, particularly in single-molecule and mass spectroscopy techniques. These approaches enable researchers to determine the distribution of specific binding stoichiometries at any ligand concentration without requiring prior information about the mechanism of ligand interaction [63].
The fundamental principle involves extracting the value of the nonspecific binding constant from the ratio of intensities corresponding to binding numbers that exceed the known number of specific binding sites (Ns). For a protein with two specific binding sites, the nonspecific association binding constant (Kn) can be determined from the intensity ratio of populations with three and four bound ligand molecules (I4/I3) using the formula:
I4/I3 = K_n[S]
where [S] represents the free ligand concentration [63]. Once the nonspecific binding constant is established, specific binding constants can be derived from other peaks in the spectra. The ratio of intensities between populations with one and zero bound ligand molecules (I1/I0) reveals the contribution of specific binding through the relationship:
I1/I0 = Kn[S] + K1[S]
enabling calculation of K1[S] = (I1 - I0Kn[S])/I_0 [63].
This mathematical framework allows researchers to correct for artificial intensity increases due to nonspecific binding and reassemble the corrected intensities according to the number of ligand molecules bound specifically to biological binding sites. The method is particularly valuable for studying MOB-NDR interactions, as it facilitates accurate quantification of binding affinities and stoichiometries without physical separation of specific and non-specific complexes.
Experimental Strategies to Overcome NSB in MOB-NDR Research
Surface Plasmon Resonance (SPR) with Reference Subtraction
Surface plasmon resonance has emerged as a gold-standard technique for measuring binding kinetics and active concentrations without calibration curves, but its application to complex biological samples requires sophisticated NSB management strategies. A particularly effective approach for studying anti-HLA antibodies (with parallels to MOB-NDR interactions) involves using a closely related non-cognate target as a reference surface [61].
The methodology entails capturing on the same flow cell first a non-cognate target and then the target of interest in a new binding cycle, carefully matching the capture levels to ensure similar NSB contributions in both situations [61]. The reference surface must be structurally similar to the specific target to mimic NSB characteristics while lacking the specific binding sites for the analyte of interest. For MOB-NDR interaction studies, this might involve using mutated versions of NDR kinases that lack MOB binding capability.
The experimental workflow consists of:
- Surface Preparation: Immobilize capture antibody (e.g., anti-B2m for HLA studies) on both flow cells
- Reference Cycle: Capture non-cognate target on the first flow cell
- Analyte Injection: Inject sample and measure response on reference surface
- Specific Cycle: In a new binding cycle, capture target of interest on the same flow cell
- Second Analyte Injection: Inject identical sample and measure total response
- Specific Signal Calculation: Subtract reference response from total response [61]
This approach successfully determined active concentrations and binding constants of anti-HLA antibodies from patients' sera, even at concentrations as low as 0.5-1 nM, despite strong NSB interference [61]. The robustness of the assay was demonstrated using a wide range of artificially generated NSB and varying densities of captured targets, confirming its applicability to challenging interaction studies.
Optimization of Experimental Conditions to Minimize NSB
Strategic optimization of experimental conditions represents a fundamental approach to reducing NSB across various methodologies. The composition of solutions profoundly impacts NSB and can be systematically optimized through several parameters:
pH Adjustment: Modifying pH to influence the dissociation state of compounds, potentially reducing ionic interactions with surfaces. The optimal pH depends on the isoelectric points of both interaction partners and should be determined empirically [62].
Buffer Composition: Incorporating buffer salts that compete with analytes for binding sites on solid surfaces. Different salt types and concentrations should be evaluated for their NSB-reduction potential without disrupting specific interactions [62].
Additive Incorporation: Introducing proteins (e.g., BSA) or surfactants that physically adsorb onto solid surfaces, blocking NSB sites. Concentration should be optimized to maximize NSB reduction while maintaining specific binding signals [62].
For material-related NSB, several strategies prove effective:
Surface Material Selection: Choosing low-adsorption consumables specifically designed to minimize molecular adherence. Different materials may be optimal for various analyte types [62].
Surface Modification: Employing chemical treatments to modify surface functional groups, reducing their interaction with analytes. Silanization of glass surfaces or plasma treatment of plastics can significantly alter binding characteristics [62].
Chromatographic System Optimization: Addressing adsorption in chromatography systems through adjustment of mobile phase ionic strength, column temperature optimization, use of inert tubing, or selection of alternative stationary phases with weaker hydrophobic interactions [62].
Table 1: Strategies for Managing Material-Related NSB
| Material Type | Primary NSB Mechanism | Recommended Solutions |
|---|---|---|
| Glassware | Ionic interactions with silanol groups | Silanization, additive incorporation |
| Polypropylene/Polystyrene | Hydrophobic interactions | Low-binding variants, surfactant addition |
| Metal Surfaces | Ionic interactions with metal cations | Passivation, mobile phase adjustment |
| Chromatography Systems | Mixed mechanisms | Inert tubing, temperature optimization |
Sample Preparation Techniques to Reduce NSB
Effective sample preparation significantly reduces NSB contributions from complex biological matrices. For serum samples prone to NSB, treatments such as dialysis or IgG purification provide partial reduction, though often insufficient for complete elimination [61]. More comprehensive approaches include:
Affinity Purification: Isulating specific analytes from complex mixtures using immobilized binding partners, significantly reducing non-specific components. For MOB-NDR studies, this could involve using immobilized MOB proteins to purify specific NDR kinases from cell lysates.
Fractionation: Separating complex samples into distinct fractions based on physicochemical properties, reducing competing components that contribute to NSB. Techniques include size-exclusion chromatography, ion-exchange chromatography, or precipitation methods.
Desalting/Buffer Exchange: Transferring samples into optimized buffers that minimize NSB while maintaining biological activity, effectively removing interfering salts or solution components.
When processing tissue samples, additional considerations are necessary, as analytes can bind to cell membrane surfaces, tissue proteins, and other components in homogenate matrices [62]. This binding can result in differential extraction recovery, particularly lower recovery at low concentrations, mimicking instability. Addressing this requires homogenization in appropriate buffers containing surfactants or competing proteins, followed by clarification steps to remove particulate matter that contributes to NSB.
Quantitative Comparison of MOB1 and MOB2 Binding Specificity
The distinct binding behaviors of MOB1 and MOB2 toward NDR kinases provide a compelling model system for studying specific protein-protein interactions. Systematic comparison of their interaction profiles reveals both quantitative and qualitative differences that underscore the importance of controlling for NSB in accurate characterization.
Table 2: Comparative Analysis of MOB1 and MOB2 Binding Properties
| Parameter | MOB1 | MOB2 |
|---|---|---|
| NDR Kinase Binding | Binds NDR1/2 with activation | Binds NDR1/2 with inhibition |
| LATS Kinase Binding | Interacts with LATS kinases | No interaction with LATS kinases |
| Functional Outcome | Increased NDR kinase activity | Diminished NDR kinase activity |
| Competitive Relationship | Competes with MOB2 for NDR binding | Competes with MOB1 for NDR binding |
| Biological Roles | Hippo signaling, cell cycle regulation | DNA damage response, cell cycle checkpoints |
MOB2's binding specificity for NDR kinases, without interaction with LATS kinases, highlights its unique role in cellular regulation [4]. Biochemical experiments demonstrate that MOB2 competes with MOB1 for NDR binding, suggesting that the relative concentrations and binding affinities of these MOB proteins determine NDR kinase activity states [4]. This competitive relationship necessitates precise quantification free from NSB artifacts.
The functional consequences of these specific interactions are biologically significant. MOB1/NDR complexes correlate with increased NDR kinase activity, while MOB2/NDR complexes associate with diminished NDR activity [4]. Furthermore, MOB2 depletion triggers p53/p21-dependent G1/S cell cycle arrest, a response not observed upon NDR1/2 knockdown, suggesting MOB2 functions in cell cycle regulation independently of NDR1/2 kinase signaling [4]. These functional differences underscore the importance of accurate, NSB-corrected binding measurements for understanding distinct biological roles.
Research Reagent Solutions for MOB-NDR Interaction Studies
Table 3: Essential Research Reagents for MOB-NDR Binding Studies
| Reagent / Material | Function / Application | Key Considerations |
|---|---|---|
| Low-Binding Consumables | Minimize analyte loss to container walls | Essential for hydrophobic compounds; various materials available |
| Surface Plasmon Resonance Chips | Immobilize binding partners for kinetic studies | CMS Series S chips commonly used with carboxymethyl dextran matrix |
| Capture Antibodies | Anchor specific targets in pull-down or SPR | Anti-B2m useful for class I HLA capture; protein-specific for NDR |
| Reference Proteins | Create NSB control surfaces | Structurally similar non-cognate targets (e.g., mutated NDR) |
| Chromatography Columns | Separate bound and unbound species | Inert surfaces minimize analyte adsorption; various chemistries |
| Buffer Additives | Reduce NSB through competition | BSA, surfactants, specific salts; concentration optimization required |
| Protease Inhibitors | Maintain protein integrity during assays | Cocktails prevent degradation during lengthy binding experiments |
Signaling Pathway Visualization
MOB Protein Regulation of NDR and DNA Damage Pathways
This diagram illustrates the dual roles of MOB proteins in cellular signaling, highlighting MOB2's involvement in DNA damage response through interaction with the MRN complex, alongside the competitive relationship between MOB1 and MOB2 in regulating NDR kinase activity.
Experimental Workflow for NSB-Managed Binding Studies
NSB-Managed Protein Interaction Workflow
This workflow outlines the key steps in conducting binding studies while accounting for non-specific binding, incorporating both general strategies and SPR-specific methodologies that utilize reference surfaces for accurate signal correction.
Overcoming non-specific binding represents a critical challenge in accurately characterizing the specific interactions between MOB proteins and NDR kinases. The distinct binding profiles of MOB1 and MOB2, with MOB2 exhibiting exclusive specificity for NDR kinases, provides an excellent model system for developing and refining NSB mitigation strategies. Through integrated approaches combining mathematical correction models, reference surface methodologies in SPR, optimized experimental conditions, and careful reagent selection, researchers can achieve the precision necessary to delineate the nuanced competitive relationship between MOB1 and MOB2 in regulating NDR kinase activity. These advanced techniques enable accurate measurement of binding parameters despite the complex biological contexts in which these interactions occur, ultimately supporting deeper understanding of their roles in cell cycle regulation, DNA damage response, and Hippo signaling pathway modulation.
Engineering Stabilized MOB Variants for Biochemical Assays
The Mps one binder (MOB) proteins are highly conserved eukaryotic cofactors essential for the function of NDR/LATS kinases, core components of Hippo signaling pathways which control cell proliferation, morphogenesis, and apoptosis [1] [4]. Mammalian genomes encode at least six different MOB proteins, with MOB1 and MOB2 exhibiting distinct binding specificities: MOB1 activates LATS kinases, while MOB2 specifically binds and regulates NDR1/2 kinases [4] [9]. This specificity is not merely structural but functional, as MOB1/NDR complexes demonstrate increased kinase activity, while MOB2/NDR complexes are associated with diminished NDR activity [4]. Engineering stabilized MOB variants is therefore a critical prerequisite for biochemical and structural studies aimed at understanding the mechanistic basis of this specificity and its implications for drug development.
Research into MOB-NDR/LATS interactions faces significant technical challenges, primarily the inherent instability of recombinant MOB proteins in heterologous expression systems [1]. This article provides a comprehensive technical guide for overcoming these challenges through the rational design of stabilized MOB variants, detailing experimental protocols for their characterization, and placing these methodologies within the broader context of MOB1 versus MOB2 binding specificity research.
Structural Basis of MOB-Kinase Interactions and Specificity
Conserved Architecture of MOB-NDR/LATS Complexes
Structural studies reveal that all NDR/LATS kinases contain a characteristic N-terminal regulatory (NTR) region that binds a specific MOB cofactor [1]. This NTR forms a V-shaped helical hairpin that interfaces with the MOB protein. The overall architecture is largely conserved, with the MOB-organized NTR mediating association of the kinase's C-terminal hydrophobic motif (HM) with an allosteric site on the N-terminal kinase lobe, representing a distinctive kinase regulation mechanism [1].
Structural Basis of MOB-NDR/LATS Interaction
Determinants of MOB1 versus MOB2 Specificity
Despite structural conservation, precise molecular recognition enables striking specificity. LATS kinases associate specifically with Mob1 proteins, while Ndr kinases associate with Mob2 proteins [1]. This specificity is restricted by discrete sites rather than being broadly distributed. Research on Saccharomyces cerevisiae Cbk1NTR-Mob2 and Dbf2NTR-Mob1 complexes identified that a short motif in the Mob structure differing between Mob1 and Mob2 strongly contributes to molecular recognition [1]. Alteration of residues in the Cbk1 NTR allows association of the noncognate Mob cofactor, confirming that specificity is mediated by limited interfacial determinants.
The biological significance of this specificity is profound. In mammalian cells, MOB2 competes with MOB1 for NDR binding, creating a potential regulatory switch [4]. The MOB1/NDR complex corresponds to increased NDR kinase activity, while the MOB2/NDR complex is associated with diminished NDR activity [4]. This competitive interaction adds a layer of regulatory complexity to Hippo signaling with implications for cell cycle progression and DNA damage response [4].
Engineering Stabilized MOB Variants: Principles and Strategies
The Stability Challenge in MOB Biochemistry
A significant obstacle in MOB kinase research is the poor stability of recombinant MOB proteins in heterologous expression systems like E. coli [1]. This instability hampers biochemical assays, structural studies, and drug discovery efforts. Researchers have noted difficulty in stably expressing monomeric Mob2 in E. coli overexpression systems, limiting progress in understanding the mechanistic basis of kinase-cofactor interactions [1].
Rational Design of Zinc-Binding MOB Variants
A successful strategy for stabilizing MOB proteins involves engineering artificial zinc-binding sites. This approach recapitulates structural motifs found naturally in metazoan orthologs [1]. For example, introducing a V148C Y153C double mutation in S. cerevisiae Mob2 creates a "zinc-binding Mob2" variant that incorporates a zinc-binding motif similar to that found in S. cerevisiae Mob1 [1].
This engineered variant demonstrates markedly improved stability while maintaining biological activity, enabling successful E. coli expression suitable for biochemistry and crystallography [1]. The strategy capitalizes on the natural structural role of zinc ions in stabilizing protein folds without disrupting functional interfaces.
Table 1: Strategies for Engineering Stabilized MOB Variants
| Strategy | Rationale | Implementation Example | Key Advantages |
|---|---|---|---|
| Zinc-Binding Site Engineering | Recapitulates natural structural motifs from metazoan orthologs | Mob2 V148C Y153C double mutation | Dramatically improves stability while maintaining function; enables crystallography |
| Discrete Specificity Determinant Optimization | Targets limited residues governing binding specificity | Mutagenesis of short recognition motif in Mob structure | Permits tuning of binding affinity without disrupting overall structure |
| NTR Complex Co-expression | Maintains natural stabilizing protein-protein interactions | Co-expression with cognate kinase NTR domain | Stabilizes native conformation; useful for complex structural studies |
Experimental Protocols for MOB Variant Characterization
Protocol 1: Engineering and Expressing Zinc-Binding MOB2
This protocol details the creation and expression of stabilized zinc-binding MOB2 variants based on published methodologies [1].
Materials:
- Wild-type MOB2 cDNA
- Site-directed mutagenesis kit
- E. coli expression strain (BL21-DE3 recommended)
- LB medium with appropriate antibiotics
- Zinc sulfate supplement (100 µM)
- Lysis buffer: 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 5% glycerol, 1 mM DTT
Method:
- Design zinc-binding variant: Introduce cysteine mutations at positions V148C and Y153C (S. cerevisiae Mob2 numbering) to create an artificial zinc-binding motif.
- Verify construct: Sequence entire ORF to confirm mutations and rule of spurious variations.
- Transform expression strain: Use BL21(DE3) E. coli with standard heat shock protocol.
- Express protein: Grow culture in LB medium at 37°C to OD600 = 0.6-0.8. Induce with 0.5 mM IPTG at 18°C for 16-18 hours. Supplement with 100 µM zinc sulfate 30 minutes pre-induction.
- Harvest and lyse cells: Pellet cells by centrifugation (4,000 × g, 20 minutes). Resuspend in lysis buffer. Lyse by sonication or French press.
- Purify protein: Use immobilized metal affinity chromatography (IMAC) followed by size exclusion chromatography. Monitor zinc incorporation by atomic absorption spectroscopy.
Protocol 2: Quantitative Binding Affinity Measurement
Surface plasmon resonance (SPR) provides quantitative data on MOB-kinase binding affinity and kinetics.
Materials:
- Biacore or similar SPR instrument
- CMS sensor chips
- Amine coupling kit
- Running buffer: HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4)
- Purified kinase NTR domain
- Stabilized MOB variants
Method:
- Immobilize ligand: Dilute kinase NTR domain to 10 µg/mL in 10 mM sodium acetate (pH 5.0). immobilize on CMS chip using standard amine coupling to achieve 500-1000 response units.
- Analyte binding: Inject stabilized MOB variants at concentrations ranging from 10 nM to 10 µM at 30 µL/min for 120-second association phase.
- Dissociation monitoring: Monitor dissociation in running buffer for 300 seconds.
- Regenerate surface: Use 10 mM glycine (pH 2.5) for 30 seconds between cycles.
- Data analysis: Subtract reference cell responses. Fit data to 1:1 Langmuir binding model to determine KD, ka, and kd.
Protocol 3: Crystallization of MOB-Kinase Complexes
This protocol enabled determination of the Cbk1NTR-Mob2 structure at 2.8 Å resolution [1].
Materials:
- Purified MOB-kinase NTR complex
- Crystallization screens (Hampton Research)
- Sitting drop vapor diffusion plates
- Cryoprotectant solutions
Method:
- Complex preparation: Mix stabilized MOB variant with kinase NTR domain in 1:1.2 molar ratio. Incubate 30 minutes on ice.
- Final purification: Apply complex to size exclusion column equilibrated with 20 mM HEPES (pH 7.5), 150 mM NaCl, 2 mM DTT.
- Concentration: Concentrate complex to 8-12 mg/mL using appropriate centrifugal concentrator.
- Crystallization screening: Set up 96-well sitting drop trials with commercial screens. Use 200 nL protein + 200 nL reservoir solution.
- Optimization: Optimize initial hits using additive screens and varying pH conditions.
- Cryoprotection and freezing: Soak crystals in cryoprotectant (reservoir solution + 25% glycerol) before flash-freezing in liquid nitrogen.
Research Reagent Solutions
Table 2: Essential Research Reagents for MOB-Kinase Studies
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Expression Systems | E. coli BL21(DE3) | Heterologous protein expression for biochemical assays |
| Stabilized MOB Variants | Zinc-binding Mob2 (V148C Y153C) | Enables structural studies and quantitative biochemistry |
| Kinase Constructs | NDR1/2 NTR domains (aa 251-351 in Cbk1) | Mapping interaction interfaces and binding specificity |
| Crystallization Tools | Commercial sparse matrix screens | Determining atomic structures of MOB-kinase complexes |
| Binding Assay Platforms | Surface plasmon resonance (Biacore) | Quantitative measurement of binding affinity and kinetics |
| Specificity Mutants | NTR with altered specificity residues | Probing determinants of MOB1 vs. MOB2 binding preference |
Data Presentation and Analysis in MOB Research
Quantitative Analysis of MOB-Kinase Interactions
Robust quantitative analysis is essential for comparing wild-type and engineered MOB variants. The following table compiles structural and binding data from key studies in this field.
Table 3: Quantitative Analysis of MOB-Kinase Complexes
| Complex | Resolution (Å) | Space Group | Binding Affinity (KD) | Key Structural Features |
|---|---|---|---|---|
| Cbk1NTR-Mob2 | 2.8 | P4₁212 | Not reported | V-shaped NTR helical hairpin; organized HM binding site |
| Dbf2NTR-Mob1 | 3.5 | P6₁222 | Not reported | Similar overall architecture with distinct specificity determinants |
| Human Lats1NTR-Mob1 | Reference [1] | Reference [1] | ~5-10 nM [3] | Conserved interface with mammalian-specific adaptations |
| Zinc-binding Mob2-Cbk1 | 2.8 [1] | P4₁212 [1] | Comparable to wild-type [1] | Enhanced stability without functional disruption |
Experimental Workflow for Stabilized MOB Characterization
Comprehensive Workflow for MOB Variant Characterization
Implications for Drug Discovery and Therapeutic Development
The structural and mechanistic insights gained from studies of stabilized MOB variants have significant implications for therapeutic development. As NDR kinases are implicated in processes ranging from cell cycle regulation and DNA damage response to microglial function in diabetic retinopathy [64] [4], targeting their regulation by MOB cofactors represents a promising therapeutic strategy.
Engineering stabilized MOB variants enables high-throughput screening for small molecules that modulate MOB-kinase interactions. Such compounds could potentially redirect signaling outcomes in pathological conditions, such as by promoting MOB2-NDR interactions to dampen kinase activity in cancer cells or enhancing MOB1-LATS interactions to activate tumor-suppressive Hippo signaling [9]. The specificity determinants identified through these structural studies provide precise molecular targets for rational drug design aimed at selectively modulating specific branches of the Hippo signaling network.
Furthermore, the demonstrated upregulation of NDR2 under high-glucose conditions in microglial cells [64] suggests potential therapeutic applications in metabolic diseases. Stabilized MOB variants could serve as tools to dissect these disease-relevant mechanisms and develop targeted interventions. As research continues to elucidate the complex roles of MOB proteins in cellular physiology, the methodologies described herein will remain essential for translating structural insights into therapeutic opportunities.
Addressing Functional Redundancy Between NDR1 and NDR2
The Nuclear Dbf2-related (NDR) kinases, NDR1 (STK38) and NDR2 (STK38L), constitute a critical subgroup of the AGC family of serine/threonine kinases with essential yet incompletely differentiated roles in cellular signaling. Despite their significant sequence homology (~86% identity) and overlapping regulatory mechanisms, emerging evidence reveals distinct, non-redundant physiological functions [65] [8]. This functional divergence is particularly evident in their interactions with Mps one binder (MOB) proteins, coregulators that dictate kinase activity and pathway specificity. Within the broader context of MOB1 versus MOB2 binding specificity research, understanding the molecular basis of NDR1/NDR2 functional redundancy represents a fundamental challenge with substantial implications for developmental biology, neurobiology, and cancer research [66] [4]. This technical guide comprehensively addresses the experimental paradigms and mechanistic insights essential for delineating unique versus overlapping functions of these paralogous kinases, providing a structured framework for researchers investigating this complex signaling nexus.
Structural and Regulatory Context of NDR Kinases
Conserved Activation Mechanism
NDR kinases operate within an evolutionarily conserved regulatory framework centered on phosphorylation events and cofactor binding. Both NDR1 and NDR2 require:
- Hydrophobic motif (HM) phosphorylation (Thr444 in NDR1, Thr442 in NDR2) by upstream MST1/2/3 kinases [66]
- Autophosphorylation of a serine residue in the activation loop (T-loop; Ser281 in NDR1, Ser282 in NDR2) [65]
- Binding of MOB coactivators (MOB1A/B) to the N-terminal regulatory (NTR) domain to relieve autoinhibition [33]
This conserved activation mechanism enables both kinases to phosphorylate downstream substrates controlling processes ranging from dendrite morphogenesis to cell cycle progression [65] [9].
Structural Determinants of MOB Binding Specificity
The interaction between NDR kinases and MOB proteins represents a critical node of pathway regulation. Structural analyses reveal that MOB1 exists in an autoinhibited state wherein its N-terminal "Switch helix" blocks the LATS1/NDR kinase-binding surface [33]. Phosphorylation of MOB1 at Thr12 and Thr35 by MST1/2 kinases induces a conformational change that relieves this autoinhibition, enabling high-affinity binding to the NTR domains of NDR1/2 and LATS1/2 kinases [33].
While MOB1 activates both NDR1 and NDR2, MOB2 exhibits distinct binding properties, competitively inhibiting NDR1/2 activation by forming a complex that displaces MOB1 [4]. This MOB2-NDR interaction is associated with diminished NDR kinase activity, creating a regulatory switch that fine-tunes NDR signaling outputs [4]. The structural basis for this differential regulation involves specific interfaces within the NTR domain that display subtle variations between NDR1 and NDR2, potentially contributing to their functional differentiation.
Figure 1: NDR Kinase Regulatory Circuitry. This diagram illustrates the core activation mechanism of NDR1/2 kinases, highlighting the opposing roles of MOB1 (activator) and MOB2 (inhibitor). MST kinase phosphorylates and activates MOB1, which then binds NDR kinases to promote their activation. MOB2 competes with MOB1 for NDR binding, forming an inhibitory complex.
Quantitative Comparison of NDR1 and NDR2 Functions
Despite their high sequence similarity, NDR1 and NDR2 display distinct expression patterns, physiological functions, and pathological associations. The following table synthesizes key comparative data extracted from functional studies.
Table 1: Functional Comparison of NDR1 and NDR2 Kinases
| Parameter | NDR1 (STK38) | NDR2 (STK38L) | Experimental Evidence |
|---|---|---|---|
| Expression in Brain | Present throughout development (P5-P20) [65] | Present throughout development (P5-P20) [65] | Western blot, immunocytochemistry [65] |
| Dendrite Morphogenesis | Limits dendrite length & proximal branching [65] | Limits dendrite length & proximal branching [65] | Dominant negative/active mutants in hippocampal neurons [65] |
| Spine/Synapse Development | Required for mushroom spine formation [65] | Required for mushroom spine formation [65] | Spine morphology analysis, mEPSC recordings [65] |
| Cancer Association | Tumor suppressor [65] [8] | Oncogene in most cancers [8] | Knockout mouse models, human cancer studies [65] [8] |
| Specific Cellular Functions | Centrosome duplication, mitosis [66] | Vesicle trafficking, autophagy, ciliogenesis [8] [66] | Substrate identification, phenotypic analysis [8] [66] |
| MOB Binding Preference | Binds both MOB1 (activating) and MOB2 (inhibitory) [4] | Binds both MOB1 (activating) and MOB2 (inhibitory) [4] | Co-immunoprecipitation, kinase assays [4] |
The phenotypic consequences of NDR perturbation further highlight both overlapping and distinct functions. The table below summarizes key findings from loss-of-function studies across different biological contexts.
Table 2: Phenotypic Outcomes of NDR Kinase Perturbation
| Organism/System | Experimental Manipulation | Observed Phenotype | Implications for Redundancy |
|---|---|---|---|
| Mammalian neurons [65] | Dominant negative NDR1/2 | Increased dendrite length & proximal branching | Functional redundancy in dendrite patterning |
| Mammalian neurons [65] | Constitutively active NDR1/2 | Reduced dendrite branching | Functional redundancy in dendrite patterning |
| C. elegans (SAX-1/NDR) [17] | sax-1/shγ87 mutation | Defective pruning of secondary/tertiary dendrite branches | Conservation of NDR function in dendrite remodeling |
| Mouse embryo [66] | Ndr1/2 double knockout | Defective somitogenesis, cardiac looping, embryonic lethality (~E10) | Essential developmental functions with partial redundancy |
| Mouse model [65] | NDR1 knockout only | Increased tumor susceptibility, NDR2 upregulation | Compensatory capacity of NDR2 for NDR1 loss |
| Human cells [4] | MOB2 knockdown | G1/S cell cycle arrest, DNA damage accumulation | MOB2-NDR interaction important for cell cycle regulation |
Experimental Approaches for Delineating Redundancy
Genetic Dissection Strategies
Compound genetic manipulations provide the most definitive evidence for addressing functional redundancy. The embryonic lethality observed in Ndr1/2 double knockout mice (~E10) contrasted with the viability of single knockout models demonstrates both essential overlapping functions and the capacity for partial compensation [66]. Tissue-specific and inducible genetic models are particularly valuable for bypassing developmental requirements and assessing post-developmental functions.
Cross-species complementation assays further illuminate functional conservation and specialization. The ability of human NDR1 to rescue dendrite patterning defects in Drosophila Tricornered (Trc) mutants indicates remarkable evolutionary conservation of core NDR functions [66]. However, the failure of specific NDR2 isoforms to fully complement all Trc functions suggests subfunctionalization has occurred during vertebrate evolution.
Biochemical Profiling Techniques
Chemical genetic substrate identification represents a powerful approach for mapping NDR kinase signaling networks. By engineering analog-sensitive kinase alleles (e.g., NDR1-as) that uniquely utilize bulky ATP analogs, researchers can specifically label and identify direct phosphorylation targets in complex biological samples [65]. This approach identified AAK1 and Rabin8 as bona fide NDR1 substrates in mouse brain, establishing concrete downstream effector pathways [65].
Comparative interactome analysis through affinity purification-mass spectrometry systematically characterizes protein-protein interaction networks for both NDR1 and NDR2 under identical experimental conditions. unpublished proteomic comparisons of NDR1 versus NDR2 interactomes in human bronchial epithelial cells (HBEC-3) and lung adenocarcinoma cells (H2030) reveal distinct interaction partners that may underlie kinase-specific functions in physiological and tumor contexts [8].
Functional Validation in Disease Contexts
The opposing roles of NDR1 and NDR2 in tumorigenesis provide a compelling context for functional dissection. While NDR1 acts as a tumor suppressor, NDR2 frequently exhibits oncogenic properties, particularly in lung cancer [8]. This divergence enables researchers to investigate how highly similar kinases acquire distinct functions in pathological settings, potentially through differential regulation of processes such as vesicle trafficking, autophagy, and immune modulation [8].
Detailed Experimental Protocols
Kinase Activity Assay for NDR1/2 Function
Purpose: To quantitatively measure NDR1 and NDR2 kinase activity in response to MOB1 versus MOB2 binding.
Reagents Required:
- Purified recombinant NDR1 or NDR2 kinase domains
- Active MOB1 and MOB2 proteins (phosphorylated or phosphomimetic)
- ATP and [γ-³²P]ATP or ADP-Glo Kinase Assay System
- NDR substrate peptide (e.g., derived from established substrates like Rabin8)
Procedure:
- Pre-incubation: Combine NDR kinase (100 nM) with either MOB1 (200 nM) or MOB2 (200 nM) in kinase assay buffer (25 mM Tris-HCl pH 7.5, 5 mM β-glycerophosphate, 2 mM DTT, 0.1 mM Na₃VO₄, 10 mM MgCl₂) for 30 minutes at 4°C.
- Reaction initiation: Add substrate peptide (200 μM) and ATP mixture (100 μM ATP + tracer [γ-³²P]ATP) to start phosphorylation reaction.
- Incubation: Maintain reactions at 30°C for 30 minutes.
- Termination and detection: Apply reaction mixture to P81 phosphocellulose paper, wash extensively with 0.75% phosphoric acid, and quantify incorporated radioactivity by scintillation counting. Alternatively, use ADP-Glo assay following manufacturer's protocol.
- Data analysis: Calculate kinase activity as pmol phosphate incorporated/min/mg kinase. Compare MOB1-stimulated versus MOB2-inhibited activities for both NDR1 and NDR2.
Technical notes: Include controls without kinase (background) and without substrate (autophosphorylation). Perform experiments in triplicate with appropriate statistical analysis. For NDR2-specific assessment, include Rabin8 as a preferential substrate [8].
Neurite Morphometry in Cultured Hippocampal Neurons
Purpose: To assess functional redundancy in dendrite patterning using RNAi and mutant overexpression.
Reagents Required:
- Dissociated hippocampal neurons from E18-E19 rat embryos
- siRNA targeting NDR1, NDR2, or non-targeting control
- Plasmids encoding GFP, kinase-dead (K118A), and constitutively active NDR1/2 mutants
- Lipofectamine 2000 or similar transfection reagent
- Immunocytochemistry reagents (anti-MAP2, anti-GFP antibodies)
Procedure:
- Culture preparation: Plate hippocampal neurons on poly-D-lysine-coated coverslips at appropriate density (50,000-70,000 cells/cm²) in Neurobasal medium with B-27 supplement.
- Gene perturbation: At DIV6-8, transfect neurons with siRNA (50 nM) and/or expression plasmids (1-2 μg) using Lipofectamine 2000 according to optimized neuronal protocol.
- Fixation and staining: At DIV16, fix neurons with 4% PFA, permeabilize with 0.1% Triton X-100, and immunostain for MAP2 (dendrites) and GFP (transfected cells).
- Image acquisition: Capture high-resolution z-stack images of GFP-positive neurons using confocal microscopy (40× or 63× objective).
- Morphometric analysis: Use automated tracing software (Neurolucida, ImageJ with NeuronJ) to quantify total dendrite length, branch points, Sholl analysis, and spine morphology.
- Statistical comparison: Compare morphological parameters between experimental groups using ANOVA with post-hoc testing.
Expected outcomes: NDR1/2 loss-of-function should increase proximal branching, while gain-of-function should restrict dendrite complexity [65]. Differential effects may emerge in spine maturation parameters.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Investigating NDR1/NDR2 Redundancy
| Reagent Category | Specific Examples | Function/Application | Key Characteristics |
|---|---|---|---|
| Chemical Inhibitors | Okadaic acid | PP2A inhibitor, indirectly activates NDR1/2 [65] | Useful for studying NDR regulation but lacks specificity |
| Activation State Markers | Anti-pT444/T442 NDR1/2 antibodies | Detect activated NDR kinases [66] | Requires validation for specific NDR isoforms |
| Genetic Tools | NDR1-KD (K118A), NDR1-AA (S281A/T444A) [65] | Kinase-dead dominant negative mutants | Block endogenous NDR function; assess necessity |
| Genetic Tools | NDR1-CA (PIFtide chimera) [65] | Constitutively active mutant | Assess sufficiency; may bypass upstream regulation |
| Substrate Reporters | AAK1, Rabin8 phosphorylation assays [65] | Direct NDR1/2 substrates | Validate kinase activity in specific pathways |
| Interaction Probes | MOB1/MOB2 binding mutants [33] [4] | Disrupt specific NDR-MOB interactions | Elucidate contribution of different MOB complexes |
| Animal Models | Ndr1/2 conditional knockout mice [66] | Tissue-specific functional analysis | Address embryonic lethality of complete knockout |
Signaling Pathways and Experimental Workflows
The molecular relationships between NDR kinases, their regulators, and effectors can be visualized through the following comprehensive pathway diagram:
Figure 2: Integrated NDR Signaling Network. This comprehensive pathway illustrates the regulatory relationships between upstream activators (MST kinases), coregulators (MOB1/2), NDR kinases, and their downstream effectors. NDR1 and NDR2 share common regulators and substrates but may exert distinct biological effects through differential expression, subcellular localization, or context-dependent interactions.
Addressing functional redundancy between NDR1 and NDR2 requires a multifaceted experimental approach that integrates genetic, biochemical, and cell biological methodologies. While these kinases undoubtedly share overlapping functions in processes such as dendrite morphogenesis and Hippo signaling, their divergent roles in tumorigenesis and specific cellular processes highlight the importance of context-dependent regulation. The differential interactions with MOB proteins, particularly the opposing effects of MOB1 (activation) versus MOB2 (inhibition), provide a critical regulatory mechanism that fine-tunes NDR kinase signaling outputs.
Future research should prioritize the development of isoform-specific chemical probes, comprehensive characterization of NDR1 versus NDR2 interactomes across different tissue contexts, and sophisticated genetic models that enable spatial and temporal control of kinase activity. Such approaches will not only resolve outstanding questions regarding NDR kinase redundancy but also illuminate their potential as therapeutic targets in cancer, neurological disorders, and developmental diseases.
Optimizing Conditions for Preserving Phosphorylation-Dependent Interactions
Protein phosphorylation represents a fundamental regulatory mechanism that controls virtually every cellular process, from signaling and metabolism to growth and apoptosis. This post-translational modification functions as a molecular switch, where the addition or removal of a phosphate group can dramatically alter protein conformation, activity, and interaction networks. Understanding and preserving these phosphorylation-dependent interactions is therefore paramount for accurate biochemical and cell biological research, particularly in the context of signaling pathways that govern cell fate and homeostasis.
This technical guide focuses specifically on the intricate relationships between MOB family adaptor proteins and their kinase binding partners, with emphasis on the Hippo tumor suppressor pathway. The MOB1 versus MOB2 binding specificity for NDR kinases presents a compelling case study for exploring phosphorylation-dependent interactions, as these interactions form a critical regulatory nexus controlling organ size, cell proliferation, and apoptosis. Research has demonstrated that MOB proteins function as central signaling nodes whose interactions are governed by precise phosphorylation events [19]. The molecular specificity of these interactions not only determines pathway output but also presents potential therapeutic targets for cancer treatment, making the optimization of their study conditions a matter of both basic and translational significance.
Molecular Foundations: MOB Proteins and NDR Kinases
The MOB Protein Family
MOB (Mps One Binder) proteins constitute an evolutionarily conserved family of adaptor proteins that function as essential co-activators of AGC group kinases. In humans, this family includes at least six members (MOB1A, MOB1B, MOB2, MOB3A, MOB3B, MOB3C, and MOB4), with MOB1A and MOB1B playing particularly critical roles in the Hippo pathway [20]. These proteins share a conserved globular core domain but differ in their N-terminal regions, which often contain regulatory elements and phosphorylation sites. MOB1 proteins are characterized by their N-terminal extension, which includes key threonine residues (Thr12 and Thr35 in mouse MOB1B) that undergo phosphorylation and regulate their activity [33].
Structural studies have revealed that full-length MOB1 exists in an autoinhibited state where the N-terminal extension, comprising a short β-strand (SN strand) and a positively-charged Switch α-helix, binds to and blocks the surface responsible for interacting with LATS/NDR kinases [33]. This autoinhibition is stabilized by β-sheet formation between the SN strand and the S2 strand of the MOB1 core domain. Phosphorylation of Thr12 and Thr35 residues structurally accelerates dissociation of the Switch helix from the LATS1-binding surface, thereby enabling LATS1 binding through a "pull-the-string" mechanism [33].
NDR Kinases and Their Regulatory Complexes
The Nuclear Dbf2-related (NDR) kinase family represents a subfamily of AGC kinases that include NDR1, NDR2, LATS1, and LATS2 in mammals [67]. These kinases form the core of the Hippo signaling pathway, which ensures proper organ size and normal tissue growth by coordinating cell proliferation and differentiation. The NDR kinases are characterized by their N-terminal regulatory (NTR) region, which is essential for kinase activity and serves as the binding site for MOB proteins [33].
The functional partnership between MOB proteins and NDR kinases is evolutionarily conserved from yeast to humans. When MOB1 binds to the NTR domain of LATS1/2 kinases, it induces a structural change that facilitates auto-phosphorylation of the activation loop, leading to full kinase activation [33]. Activated MOB1-bound LATS1/2 then phosphorylates downstream transcriptional co-activators YAP and TAZ, resulting in their cytoplasmic retention and degradation, thereby inhibiting their transcriptional programs promoting cell proliferation [67].
Table 1: Core Components of MOB-NDR/LATS Signaling Axis
| Component | Gene(s) | Function | Regulatory Features |
|---|---|---|---|
| MOB1A/MOB1B | MOB1A, MOB1B | Core co-activator of LATS1/2 | Activated by MST1/2 phosphorylation at Thr12/Thr35 |
| MOB2 | MOB2 | Co-activator of NDR1/2 | Phosphopeptide binding capability [19] |
| NDR1/NDR2 | STK38, STK38L | Serine/threonine kinases | Regulate cell cycle, transcription, apoptosis [67] |
| LATS1/LATS2 | LATS1, LATS2 | Tumor suppressor kinases | Phosphorylate YAP/TAZ; require MOB1 binding [33] |
| MST1/MST2 | STK4, STK3 | Upstream kinases | Phosphorylate MOB1 and LATS1/2 [19] |
Structural Basis of Phosphorylation-Dependent Interactions
MOB1 Phospho-Recognition Mechanisms
The molecular basis for phosphorylation-dependent interactions between MOB1 and its binding partners has been elucidated through structural and biochemical studies. MOB1 contains a highly conserved phospho-recognition infrastructure composed of three basic residues (Lys153, Arg154, and Arg157 in human MOB1) that form a binding pocket for phosphate moieties [19]. This infrastructure enables MOB1 to recognize and bind phosphothreonine-containing sequences in its interaction partners.
Structural analyses have revealed that MOB1's phosphopeptide-binding specificity is complementary to the substrate phosphorylation consensus of its upstream kinases MST1 and MST2 [19]. This complementarity ensures signaling fidelity within the Hippo pathway. The crystal structures of MOB1A in complex with two favored phosphopeptide sites in MST1 (pT353 and pT367) provided a comprehensive description of the MOB1A phosphopeptide-binding consensus, revealing how specific sequence contexts surrounding phosphothreonine residues influence binding affinity [19].
Interestingly, systematic examination of MOB1's phosphopeptide binding specificity demonstrated that all but one of the human MOB proteins share the ability to bind MST1 phosphopeptides, suggesting conservation of this phospho-recognition mechanism across most MOB family members [19]. This finding provides a foundation for understanding the functional relationships between different MOB proteins and their kinase partners.
MOB1 Versus MOB2 Binding Specificity
While MOB1 and MOB2 share structural similarities, they exhibit distinct binding specificities for NDR kinases. MOB1 primarily interacts with and activates LATS1/2 kinases, whereas MOB2 shows preference for NDR1/2 kinases [13]. This specificity is determined by structural differences in their binding surfaces and the distinct conformational requirements of their kinase partners.
Research has demonstrated that the phosphorylation-dependent recruitment of signaling complexes differs between MOB1 and MOB2. For instance, while MOB1's interaction with MST kinases is mediated by its phosphopeptide-binding domain, its binding to LATS and NDR kinases occurs through a distinct interaction surface [20]. This multifunctional nature allows MOB1 to simultaneously engage both upstream and downstream kinases, facilitating trans-phosphorylation events that activate the Hippo pathway.
The differential binding specificities of MOB1 and MOB2 for NDR kinases underscore the complexity of phosphorylation-dependent interaction networks and highlight the importance of maintaining pathway specificity through precise molecular recognition mechanisms.
Quantitative Analysis of Phosphorylation-Dependent Binding
The affinity of phosphorylation-dependent interactions can be quantitatively assessed using various biophysical techniques. Studies on MOB1 interactions have revealed substantial differences in binding affinity between phosphorylated and non-phosphorylated states, with phosphorylation often increasing binding affinity by several orders of magnitude.
Table 2: Quantitative Binding Data for Phosphorylation-Dependent Interactions
| Interaction Pair | Phosphorylation Status | Affinity/Binding Strength | Method | Reference |
|---|---|---|---|---|
| MOB1-MST1 | Phosphorylated at Thr353 | Kd ≈ 2.4 μM for optimized peptide | FP | [19] |
| MOB1-MST2 | Phosphorylated at Thr378 | Bipartite binding mode observed | Crystallography | [19] |
| MOB1-LATS1 | MOB1 phosphorylated at Thr12/Thr35 | Essential for high-affinity binding | Biochemical assays | [33] |
| MOB1-NDR1 | MOB1-dependent activation | Rapid activation at membrane | Cellular assays | [13] |
| MOB2-NDR1/2 | Phosphorylation-dependent | Stimulates kinase activity in vitro | Kinase assays | [13] |
Fluorescence polarization binding experiments with FITC-labeled phosphorylated and non-phosphorylated MST1 peptides have provided detailed quantitative information about the binding thermodynamics and kinetics of these interactions [19]. These studies demonstrated that the phosphorylation-dependent interaction between MOB1 and MST kinases follows a characteristic binding curve with rapid association and dissociation rates, consistent with the dynamic nature of signaling interactions.
Membrane-targeted hMOBs robustly promoted activation of NDR, and in vivo activation of human NDR by membrane-bound hMOBs was found to be dependent on their interaction and occurs solely at the membrane [13]. Using a chimeric molecule of hMOB that allows inducible membrane translocation, researchers found that NDR phosphorylation and activation at the membrane occur within minutes after association of hMOB with membranous structures, highlighting the rapid kinetics of these phosphorylation-dependent interactions [13].
Essential Methodologies for Studying Phosphorylation-Dependent Interactions
Protein Expression and Purification Protocols
Recombinant Protein Expression in E. coli For structural and biophysical studies, human MOB and kinase domains are typically expressed in E. coli BL21 (DE3) CodonPlus RIL cells as N-terminal dual 6xhistidine (HIS) and glutathione S-transferase (GST) fusion proteins using modified pETM-30 vectors [19]. The tobacco etch virus (TEV) protease cleavage site between the affinity tags and the protein of interest allows for tag removal after purification. For full-length MOB1 proteins, which are prone to proteolytic degradation of their N-terminal regions, low-temperature expression and rapid purification are critical for obtaining intact protein [33].
Purification and Cleavage Proteins are purified in batch using glutathione-Sepharose resin, followed by cleavage from affinity tags with HIS-tagged TEV protease [19]. The TEV protease is subsequently removed by immobilized-metal affinity chromatography. The cleaved protein is then concentrated and subjected to size exclusion chromatography (SEC) using a Superdex 75 column for final polishing and buffer exchange. For proteins used in pull-down assays and Far-Western analysis, the TEV cleavage step may be omitted to retain the affinity tag.
Crystallization and Structural Analysis
Complex Formation and Crystallization Complexes of MOB1 with phosphopeptides are obtained by mixing at a 1:1.5 mole ratio [19]. Crystals of MOB1-phosphopeptide complexes are typically obtained by vapor diffusion using hanging drops with 1:1 mixtures of protein (at 7 mg/ml concentration) with a precipitant solution of 0.1 M MES pH 6.0, 0.2 M LiCl and 20% PEG 6000. For diffraction studies, protein crystals are flash-frozen in mother liquors supplemented with 20-25% (v/v) ethylene glycol as cryoprotectant.
Data Collection and Structure Determination X-ray diffraction data are collected at synchrotron sources, such as the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences [20]. Data processing typically involves programs like HKL-2000 or XDS, followed by structure solution using molecular replacement with existing MOB structures as search models. Iterative model building and refinement are performed using programs like Coot and Phenix.
Interaction Proteomics and Phosphopeptide Binding Studies
Interaction Proteomics To identify phosphorylation-dependent interactions, affinity purification coupled with mass spectrometry is employed [20]. This approach involves expressing bait proteins in cell lines, performing affinity purification under carefully controlled conditions, and then identifying co-purifying proteins by liquid chromatography-tandem mass spectrometry (LC-MS/MS). To preserve phosphorylation-dependent interactions, phosphatase inhibitors must be included in all lysis and wash buffers.
Phosphopeptide Binding Specificity Analysis The binding specificity of MOB proteins for phosphopeptides is systematically examined using proteomic approaches combined with peptide arrays and quantitative binding analyses [19]. Peptide arrays containing systematically varied sequences surrounding phosphorylation sites are probed with MOB proteins to determine sequence preferences. Quantitative binding analyses using fluorescence polarization with FITC-labeled peptides provide dissociation constants for different peptide sequences.
Optimization Strategies for Preserving Phosphorylation-Dependent Interactions
Sample Preparation and Handling
Preservation of Phosphorylation Status Maintaining the native phosphorylation state of proteins during sample preparation is paramount for studying phosphorylation-dependent interactions. Several strategies are essential:
Rapid Protein Denaturation: To preserve the in-vivo state of the phosphoproteome and reduce post-mortem effects, endogenous enzymatic activity should be eliminated immediately after sample collection through thermal protein denaturation using specialized equipment like the Stabilizor T1 system [68]. This procedure effectively abolishes the activity of protein phosphatases, kinases, proteases and other enzymes that can alter protein modification sites during sample handling.
Comprehensive Protease and Phosphatase Inhibition: During protein extraction and purification, include broad-spectrum protease inhibitors (e.g., PMSF, aprotinin, leupeptin) and phosphatase inhibitors (e.g., sodium fluoride, β-glycerophosphate, sodium orthovanadate) in all buffers. For particularly sensitive interactions, use more specific phosphatase inhibitor cocktails targeting serine/threonine and tyrosine phosphatases.
Controlled Extraction Conditions: Perform protein extractions at 4°C using optimized lysis buffers that maintain protein stability and interactions. Urea-based buffers have been successfully employed for tissue phosphoproteome preservation, followed by careful homogenization using ceramic beads in systems like the Precellys 24 [68].
Buffer Composition and Stabilization
The composition of buffers used throughout protein purification and analysis significantly impacts the stability of phosphorylation-dependent interactions. Key considerations include:
Ionic Strength Optimization: The interactions between MOB proteins and their kinase partners often involve charged residues and phospho-groups. Buffer ionic strength should be optimized to mimic physiological conditions (typically 150-200 mM NaCl) while minimizing non-specific interactions.
pH Stability: Maintain buffers at pH 7.0-7.5 to preserve native protein structure and interaction interfaces. The use of buffering agents such as HEPES, Tris, or phosphate buffers at appropriate concentrations (20-50 mM) provides adequate capacity during purification steps.
Reducing Environment: Include reducing agents such as dithiothreitol (DTT) or β-mercaptoethanol (1-5 mM) to prevent oxidative damage to cysteine residues, which is particularly important for MOB proteins that contain conserved cysteine residues involved in structural zinc coordination [33].
Divalent Cation Management: For MOB and NDR kinase interactions, include magnesium ions (1-5 mM MgCl₂) in buffers, as these are often required for kinase activity and phospho-dependent interactions. However, omit EDTA and EGTA unless specifically studying metal-dependent processes.
Experimental Conditions for Binding Assays
Temperature and Time Considerations Phosphorylation-dependent interactions are often dynamic and sensitive to experimental conditions:
Low-Temperature Operations: Conduct protein purification and binding assays at 4°C whenever possible to slow enzymatic degradation and preserve labile phosphorylation-dependent complexes [33].
Rapid Processing: Minimize the time between protein extraction and analysis. Studies have shown that even with inhibitors, prolonged processing can lead to significant changes in phosphorylation status.
Controlled Incubation Times: For in vitro binding assays, optimize incubation times to achieve equilibrium without allowing significant dephosphorylation. Time-course experiments should be performed to establish appropriate durations.
Stabilization Additives Include specific additives in assay buffers to stabilize phosphorylation-dependent interactions:
ATP Analogs: For kinase-substrate interactions, include non-hydrolyzable ATP analogs in binding assays to stabilize the complex without promoting phosphorylation turnover.
Phosphatase Inhibitors: Use specific phosphatase inhibitors such as okadaic acid (for PP1 and PP2A), calyculin A, or fostriecin in binding assays to prevent dephosphorylation during incubation [13].
Carrier Proteins: Include inert carrier proteins like bovine serum albumin (BSA, 0.1-1 mg/ml) to prevent non-specific binding and stabilize dilute protein solutions.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Studying Phosphorylation-Dependent Interactions
| Reagent/Category | Specific Examples | Function/Application | Optimization Notes |
|---|---|---|---|
| Phosphatase Inhibitors | Sodium orthovanadate, β-glycerophosphate, Okadaic acid | Preserve phosphorylation status during extraction | Use combinations for broad-spectrum inhibition; okadaic acid (1 μM) for PP1/PP2A [13] |
| Protease Inhibitors | PMSF, Aprotinin, Leupeptin, Pepstatin A | Prevent protein degradation | Use EDTA-free cocktails when studying metal-dependent interactions |
| Affinity Tags | HIS, GST, MBP | Protein purification and pull-down assays | Dual HIS-GST tags with TEV cleavage site provide flexibility [19] |
| Lysis Buffers | Urea buffer, RIPA buffer, NP-40 buffer | Protein extraction with maintained interactions | Urea buffer effective for tissue phosphoproteome preservation [68] |
| Crystallization Reagents | MES, LiCl, PEG 6000, ethylene glycol | Protein crystallization and cryoprotection | 0.1 M MES pH 6.0, 0.2 M LiCl, 20% PEG 6000 successful for MOB1 complexes [19] |
| Kinase Assay Components | ATP, MgCl₂, MnCl₂, kinase buffers | In vitro kinase activity measurements | Include phosphatase inhibitors in all kinase assay buffers |
Signaling Pathway Visualization
Diagram 1: Hippo Pathway Regulation by MOB1 Phosphorylation
Experimental Workflow for Interaction Studies
Diagram 2: Experimental Workflow for Preserving Phosphorylation-Dependent Interactions
The study of phosphorylation-dependent interactions between MOB proteins and NDR kinases requires meticulous attention to experimental conditions throughout the workflow, from sample collection to data analysis. The optimization strategies outlined in this guide—focusing on rapid sample processing, comprehensive enzyme inhibition, buffer optimization, and appropriate binding assay conditions—provide a foundation for reliable investigation of these critical signaling interactions.
As research in this field advances, several areas warrant particular attention. First, the development of more specific phosphatase inhibitors with minimal off-target effects would significantly improve the preservation of native phosphorylation states. Second, techniques for stabilizing transient phosphorylation-dependent complexes for structural studies continue to evolve, with crosslinking strategies and cryo-electron microscopy offering promising avenues. Finally, the integration of computational approaches with experimental data will enhance our understanding of the dynamic nature of these interactions and facilitate the prediction of binding interfaces.
The MOB1 versus MOB2 binding specificity for NDR kinases exemplifies how subtle molecular differences can translate into distinct biological outcomes through phosphorylation-dependent interactions. By applying the optimized conditions and methodologies described in this guide, researchers can advance our understanding of these crucial regulatory mechanisms and their implications for health and disease.
Navigating MOB1-MOB2 Competition for NDR Binding
The regulation of Nuclear Dbf2-related (NDR) kinases represents a critical control point in eukaryotic signaling, influencing processes from cell proliferation to DNA damage response. Central to this regulation is a competitive interaction between Mps one binder (MOB) proteins MOB1 and MOB2, which bind to the same site on NDR kinases yet exert opposing functional effects. This whitepaper synthesizes current structural and biochemical research to delineate the precise molecular mechanisms governing MOB1-MOB2 competition. We provide an in-depth analysis of how this competition integrates with the broader Hippo signaling network and present standardized experimental methodologies for investigating these protein-protein interactions. The mechanistic insights and technical frameworks presented herein aim to facilitate advanced research and therapeutic targeting of this crucial regulatory system.
NDR kinases (NDR1/2, also known as STK38/STK38L) are evolutionarily conserved serine-threonine kinases belonging to the AGC kinase family, functioning as pivotal signaling nodes in cellular processes including centrosome duplication, mitotic progression, DNA damage response, and Hippo pathway signaling [4] [69]. Their activity is stringently controlled through association with MOB coactivator proteins, which serve as essential regulatory subunits. Mammalian cells express multiple MOB proteins, with MOB1 and MOB2 demonstrating particularly significant yet antagonistic roles in NDR regulation.
MOB1 association with NDR kinases promotes kinase activation through facilitating autophosphorylation and supporting phosphorylation by upstream kinases [16] [3]. In stark contrast, MOB2 competes with MOB1 for binding to the same N-terminal regulatory (NTR) domain on NDR kinases, yet this interaction is associated with diminished NDR kinase activity [70] [16]. This competitive binding establishes a molecular switch that determines NDR activation status, thereby influencing downstream cellular responses. The MOB1-MOB2-NDR axis thus represents a critical regulatory module whose dysregulation may contribute to pathological conditions, including cancer development and progression.
Structural Basis of MOB-NDR Interactions
The NDR N-Terminal Regulatory (NTR) Domain: A Shared Binding Platform
The structural determinant for MOB protein binding is the NTR domain of NDR kinases, which adopts a V-shaped helical hairpin conformation [1] [2]. This structural motif is conserved across the NDR/LATS kinase family and creates a dedicated binding interface for MOB proteins. Biophysical and crystallographic studies have revealed that both MOB1 and MOB2 bind to this same site on the NTR, creating the structural foundation for their competitive relationship [1] [16].
Table 1: Key Structural Features of MOB-NDR Complexes
| Structural Element | Role in MOB Binding | Consequence of Binding |
|---|---|---|
| NDR NTR Domain | Forms V-shaped helical hairpin; provides binding interface | Organizes MOB cofactor; necessary for allosteric regulation |
| MOB1 Structure | Binds NTR; interacts with kinase hydrophobic motif (HM) | Promotes NDR autophosphorylation; enhances kinase activity |
| MOB2 Structure | Binds same NTR site as MOB1 via similar interface | Competes with MOB1; prevents activation; maintains inactive state |
| Activation Segment | Atypically long in NDR1; autoinhibitory | MOB1 binding partially relieves inhibition; distinct from MOB effect |
Molecular Determinants of Binding Specificity
Despite binding the same general region on the NTR, MOB1 and MOB2 exhibit distinct binding modes and specificities driven by discrete structural elements. High-resolution crystal structures of Saccharomyces cerevisiae Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes have identified a short motif within the Mob structure that differs between MOB1 and MOB2, strongly contributing to molecular recognition specificity [1]. Mutational alteration of residues in this region allows association with noncognate cofactors, indicating that specificity is restricted to discrete sites rather than distributed broadly across the interaction surface [1].
The functional outcome of MOB binding is fundamentally different: MOB1 binding organizes the NTR to interact with the AGC kinase C-terminal hydrophobic motif (HM), facilitating kinase activation [1]. In contrast, MOB2 binds preferentially to unphosphorylated NDR and appears to stabilize an inactive kinase conformation [16]. This structural understanding explains how competition at a shared site can yield divergent functional outcomes for NDR kinase activity.
Functional Consequences of Competitive Binding
Biochemical Regulation of NDR Kinase Activity
The competitive binding between MOB1 and MOB2 creates a tunable regulatory mechanism for NDR kinase activity. Biochemical studies demonstrate that MOB2 competes with MOB1 for NDR binding, with the MOB1/NDR complex corresponding to increased NDR kinase activity while the MOB2/NDR complex is associated with diminished NDR activity [4] [16]. RNA interference-mediated depletion of MOB2 results in increased NDR kinase activity, consistent with its role as a negative regulator [16].
This competition extends to the regulation of downstream effectors. Research in SMMC-7721 hepatocellular carcinoma cells demonstrates that MOB2 knockout promotes cell migration and invasion, increases NDR1/2 phosphorylation, and decreases phosphorylation of the Hippo pathway effector YAP (Yes-associated protein) [70] [23]. Conversely, MOB2 overexpression produces the opposite effects, suggesting that the MOB1-MOB2 competition influences Hippo pathway signaling through modulation of NDR kinase activity toward downstream targets including YAP [70].
Integration with Hippo Signaling and Cellular Processes
The MOB1-MOB2-NDR regulatory module integrates with broader signaling networks, particularly the Hippo tumor suppressor pathway. While LATS kinases are traditionally considered the primary YAP kinases in Hippo signaling, NDR1/2 also function as direct YAP kinases, phosphorylating YAP on multiple sites (Ser61, Ser109, Ser127, Ser164) to inhibit its transcriptional coactivator function [69]. The competition between MOB1 and MOB2 for NDR binding therefore directly influences Hippo pathway output and YAP/TAZ-mediated transcription.
Table 2: Functional Outcomes of MOB-NDR Interactions
| Cellular Process | MOB1-NDR Role | MOB2-NDR Role | Experimental Evidence |
|---|---|---|---|
| Cell Motility | Inhibits migration/invasion | Promotes migration/invasion when dominant | Wound-healing/Transwell assays in HCC cells [70] |
| Hippo Signaling | Activates LATS/YAP phosphorylation | Decreases YAP phosphorylation | Immunoblotting for pYAP in MOB2 KO/OE cells [70] |
| Cell Cycle | Supports G1/S progression | Depletion causes G1/S arrest via p53/p21 | Cell cycle analysis after MOB2 knockdown [4] |
| DNA Damage Response | Not well characterized | Required for DDR signaling and cell survival | ATM activation studies post-irradiation [4] |
Beyond Hippo signaling, the MOB1-MOB2 competition influences diverse cellular processes. MOB2 knockdown activates a p53/p21-dependent G1/S cell cycle checkpoint and causes accumulation of DNA damage, suggesting roles in cell cycle progression and genome maintenance [4]. These effects appear to be at least partially independent of NDR kinases, indicating that MOB2 may have additional binding partners beyond NDR1/2, such as the RAD50 component of the MRN DNA damage sensor complex [4].
Diagram 1: MOB1-MOB2 Competition in Hippo Signaling. MOB1 (yellow) activates NDR kinases, while MOB2 (red) competes for binding and inhibits NDR. Both NDR and LATS kinases phosphorylate YAP, leading to cytoplasmic retention and inhibited gene expression.
Experimental Approaches for Studying MOB-NDR Interactions
Structural Biology Methodologies
Protein Complex Crystallography: Structural characterization of MOB-NDR complexes requires expression and purification of the NDR NTR domain (residues 251-351 in Cbk1) and MOB proteins (residues 45-287 in Mob2) [1]. For unstable MOB proteins such as Mob2, engineering a zinc-binding motif (V148C Y153C in scMob2) can improve stability for crystallization [1]. Crystallization conditions for the Cbk1NTR–Mob2 complex included 0.1 M sodium citrate (pH 5.5) and 10% PEG 8000, with data collection at 2.8 Å resolution [1]. Structural analysis focuses on the interface between the NTR helical hairpin and the MOB protein surface, with particular attention to residues governing specificity.
Biophysical Binding Assays: Surface plasmon resonance (SPR) or fluorescence polarization (FP) assays quantitatively measure MOB-NDR binding affinity and kinetics. These techniques can characterize how mutations in the NTR or MOB proteins affect binding specificity and strength. Competition assays using these biophysical methods can directly demonstrate MOB1-MOB2 competition by measuring displacement of one MOB protein by the other [16].
Cell Biological Assessment
Lentiviral Manipulation of MOB Expression: To investigate functional consequences in cellular models, lentiviral vectors encoding MOB2 (LV-MOB2) or CRISPR/Cas9 constructs for MOB2 knockout (LV-sgMOB2) can be used to establish stable cell lines [70] [23]. For MOB2 knockout, the sgRNA sequence 5'-AGAAGCCCGCTGCGGAGGAG-3' targets the MOB2 coding sequence [70]. Following lentiviral infection, selection with 1.0 µg/ml puromycin for two weeks establishes stably transduced populations, with efficacy verified by western blotting [70].
Functional Assays for NDR Activity:
- Kinase Activity Assays: Immunoprecipitation of NDR kinases followed by in vitro kinase assays using specific substrates (e.g., YAP-derived peptides) quantifies NDR activation status under different MOB expression conditions [69].
- Wound Healing and Transwell Assays: These measure cell motility changes resulting from manipulated MOB-NDR interactions. Cells are serum-starved overnight, wounded with a pipette tip, and migration is quantified over 48 hours in 1% FBS [70].
- Immunofluorescence and Co-localization: Endogenous or tagged NDR and MOB proteins can be visualized to assess subcellular localization and co-localization under different conditions, providing insights into spatial regulation of these interactions [3].
Diagram 2: Experimental Workflow for MOB-NDR Interaction Studies. A comprehensive approach integrating structural, biochemical, and cellular methodologies to characterize MOB1-MOB2 competition and its functional consequences.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Investigating MOB-NDR Interactions
| Reagent/Tool | Specifications | Application | Key Considerations |
|---|---|---|---|
| NDR NTR Constructs | Cbk1 residues 251-351; Dbf2 NTR domain | Structural studies; binding assays | Maintains V-shaped helical hairpin; sufficient for MOB binding |
| MOB Expression Vectors | pcDNA3, pGEX-4T1, pMal-2c with MOB1/2 | Protein expression; cellular studies | MOB2 may require stabilization (zinc-binding motif) |
| CRISPR sgMOB2 | 5'-AGAAGCCCGCTGCGGAGGAG-3' | MOB2 knockout models | Verify knockout by western blot; monitor compensatory effects |
| Lentiviral MOB2 | LV-MOB2 (OE); LV-sgMOB2 (KO) | Stable cell line generation | Use 1.0 µg/ml puromycin for selection; confirm expression |
| Anti-NDR Antibodies | Phospho-specific (Ser281/282; Thr444/442) | Activity assessment | Distinguish activation states; monitor HM phosphorylation |
| YAP Substrates | Peptides with HXRXXS/T motifs | Kinase activity assays | NDR phosphorylates YAP on Ser61, Ser109, Ser127, Ser164 |
The competitive interaction between MOB1 and MOB2 for NDR binding represents a sophisticated regulatory mechanism that fine-tunes NDR kinase activity and its downstream cellular effects. Structural biology has illuminated the molecular basis of this competition, revealing how two related proteins binding to the same site can exert opposing functional effects through distinct binding modes and outcomes for kinase activation. The integration of this regulatory module with the Hippo pathway and other signaling networks underscores its biological significance in processes ranging from cell proliferation to DNA damage response.
Future research should focus on elucidating the spatial and temporal regulation of this competition within cells, particularly how upstream signals might bias the balance toward MOB1 or MOB2 binding. The development of small molecules that specifically modulate these interactions could provide both therapeutic leads and research tools for precisely manipulating this pathway. Additionally, further investigation of MOB2's NDR-independent functions, particularly its role in DNA damage response through interactions with RAD50, may reveal novel biological functions beyond its competitive relationship with MOB1. As our structural and mechanistic understanding deepens, so too will our ability to target this system for therapeutic benefit in cancer and other diseases characterized by dysregulated cell growth and survival.
Strategies for Differentiating Direct vs. Indirect Regulatory Effects
In molecular biology, accurately distinguishing direct regulatory effects from indirect ones is fundamental to understanding true mechanism of action. This challenge is particularly acute in kinase research, where signaling cascades can create complex networks of interactions. Within the context of MOB1 versus MOB2 binding specificity for NDR kinases, employing rigorous strategies to differentiate direct physical binding from indirect regulatory consequences becomes paramount for valid mechanistic interpretation. This technical guide provides researchers with experimental frameworks and analytical methodologies to dissect these relationships, leveraging advanced statistical approaches and direct binding assays to establish causal regulatory mechanisms with high confidence. The principles outlined are essential for drug development professionals targeting specific kinase pathways, where misattribution of direct effects can lead to costly development failures.
Cellular signaling networks comprise numerous interconnected pathways where distinguishing direct physical interactions from downstream consequences represents a significant experimental challenge. A direct regulatory effect occurs when a molecule physically interacts with its target to modulate activity, such as a MOB coactivator binding directly to an NDR kinase catalytic domain. In contrast, an indirect regulatory effect manifests through intermediate molecules or secondary pathways, where the observed outcome results from a cascade of events rather than physical contact. In the context of NDR kinase regulation, the highly specific binding of MOB1 to Lats kinases and MOB2 to Ndr kinases exemplifies a system where differentiating these effects is crucial for understanding pathway specificity [1].
The evolutionary conservation of Hippo signaling pathways from yeast to humans underscores the fundamental importance of these regulatory mechanisms in controlling cell proliferation and morphogenesis [1]. The development of selective therapeutic interventions requires precise mapping of these interactions, as drugs targeting direct binding interfaces offer different therapeutic potential than those affecting indirect regulators. This guide establishes a comprehensive framework for employing statistical, biochemical, and computational approaches to differentiate these regulatory modes with emphasis on MOB-NDR kinase research.
Statistical Frameworks for Comparison
Direct vs. Indirect Comparison Methodologies
In the absence of head-to-head experimental data, researchers often employ statistical methods to infer relationships between molecular entities. Naïve direct comparisons—directly comparing results from separate experiments without adjustment—are generally inappropriate for establishing direct regulatory relationships as they ignore contextual differences in experimental conditions [71]. These comparisons break the original randomization of individual experiments and introduce significant confounding and bias because systematic differences between experimental setups may be misinterpreted as true biological effects [71].
Adjusted indirect comparisons preserve randomization by comparing the effect of two interventions relative to a common comparator [71]. This method uses a connected network where Treatment A vs. Comparator C and Treatment B vs. Comparator C allows inference about A vs. B. For protein interaction studies, this could involve comparing the binding affinities of MOB1 and MOB2 to a common NDR kinase domain variant before drawing conclusions about their relative specificity.
Mixed treatment comparisons incorporate Bayesian statistical models to integrate all available data, including studies not directly relevant to the primary comparison [71]. These advanced methods reduce uncertainty but have not yet gained widespread acceptance in regulatory decision-making for drug development.
Table 1: Comparison of Statistical Methods for Establishing Regulatory Relationships
| Method | Principle | Advantages | Limitations | Suitable Experimental Context |
|---|---|---|---|---|
| Naïve Direct Comparison | Direct comparison of results from separate experiments | Simple to perform; requires only summary data | High potential for confounding; ignores differences in experimental conditions | Preliminary exploration only; when no other options exist |
| Adjusted Indirect Comparison | Comparison through common comparator using relative effects | Preserves randomization; accepted by health technology assessment agencies | Increased statistical uncertainty; requires common comparator | Comparing MOB binding affinities using a common kinase domain |
| Mixed Treatment Comparison | Bayesian network incorporating direct and indirect evidence | Reduces uncertainty; incorporates all available evidence | Computational complexity; not widely accepted by regulators | Integrating multiple binding studies across related kinase families |
Quantitative Synthesis and Reporting
When synthesizing quantitative data from multiple experiments for comparative analysis, systematic reporting is essential. The Agency for Healthcare Research and Quality recommends including descriptive study information, level of evidence assessment, and graphical presentation of individual and combined study estimates [72]. For protein interaction studies, this should encompass:
- Sample sizes and experimental conditions for each binding assay
- Quantitative measures of effect (e.g., Kd values, binding affinity fold-changes)
- Confidence intervals for all point estimates
- Assessment of statistical heterogeneity between experimental replicates
- Sensitivity analyses testing key assumptions
Forest plots should present both individual experiment results and combined estimates where appropriate, with weights assigned to each study based on precision [72]. This approach allows researchers to differentiate consistent direct effects from variable indirect effects that may depend on specific experimental conditions.
MOB-NDR Kinase Binding Specificity
Structural Determinants of Direct Binding
The specific binding between MOB coactivators and NDR/Lats kinases provides an exemplary model system for studying direct regulatory effects. Structural analyses reveal that NDR/Lats kinases contain a characteristic N-terminal regulatory (NTR) region that binds specifically to distinct MOB cofactors: Lats kinases associate with Mob1 proteins, while Ndr kinases associate with Mob2 proteins [1]. This specific recognition is remarkable given the high conservation between both the kinase NTR regions and the MOB cofactors across the entire protein family.
Crystallographic studies of Saccharomyces cerevisiae Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes demonstrate that the NTR forms a V-shaped helical hairpin that interfaces with the MOB cofactor [1]. This interaction serves as a structural platform that mediates kinase-cofactor binding specificity. The improved resolution of these crystal structures highlights the critical role of MOB binding in positioning the hydrophobic motif (HM) of the kinase, which upon phosphorylation activates the enzyme [1]. This represents a direct regulatory mechanism where MOB binding organizes the NTR to interact with the AGC kinase C-terminal hydrophobic motif, facilitating HM engagement with an allosteric site on the N-terminal kinase lobe.
Table 2: Experimental Evidence for MOB-NDR/Lats Specific Binding
| Kinase | Cofactor | Organism | Key Evidence | Functional Context |
|---|---|---|---|---|
| Cbk1 (NDR) | Mob2 | S. cerevisiae | Crystal structure of complex; specific association even with simultaneous presence of Mob1 | RAM network; cell separation and polarized growth |
| Dbf2 (Lats) | Mob1 | S. cerevisiae | Crystal structure of complex; specific association despite presence of Mob2 | Mitotic Exit Network (MEN); cytokinesis and M to G1 transition |
| NDR1 | Mob2 | H. sapiens | Co-immunoprecipitation; colocalization; dramatic kinase activation | Cell proliferation; tumor progression; cytoplasmic localization |
| NDR2 | Mob2 | H. sapiens | Co-immunoprecipitation; colocalization; dramatic kinase activation | Thymus expression; punctate cytoplasmic distribution |
Specificity Mechanisms and Functional Consequences
The specificity of MOB-NDR kinase interactions is enforced by discrete molecular recognition sites rather than broadly distributed interface properties. Mutational analyses indicate that altering specific residues in the Cbk1 NTR allows association with noncognate Mob cofactors, demonstrating that specificity is restricted to critical contact points [1]. This finding has profound implications for distinguishing direct from indirect effects, as minimal structural changes can discriminate between specific binding partners.
Functional studies demonstrate that MOB binding directly activates NDR kinases. Human NDR1 and NDR2 form stable complexes with Mob2, and this association dramatically stimulates their catalytic activity [3]. This direct regulatory effect identifies MOB proteins as a unique class of kinase-activating subunits functionally analogous to cyclins in cell cycle regulation. The differential subcellular localization of NDR1 (nuclear) and NDR2 (punctate cytoplasmic) suggests that despite sharing the same MOB cofactor, these kinases may serve distinct cellular functions through spatial segregation of their direct regulatory complexes [3].
Experimental Approaches for Differentiation
Direct Binding Assays
Establishing direct regulatory relationships requires experimental demonstration of physical interaction without intermediary components. Several key methodologies provide this evidence:
Co-immunoprecipitation under stringent conditions can demonstrate direct associations, as shown for epitope-tagged NDR kinases immunoprecipitated from Jurkat T-cells which identified interactions with MOB homologs [3]. Critical controls include testing for non-specific interactions and verifying that complex formation persists under conditions that disrupt larger protein complexes.
Surface Plasmon Resonance (SPR) provides quantitative data on binding kinetics without cellular context. Purified kinase domains and MOB cofactors should demonstrate direct binding with characteristic association and dissociation rates. The concentration dependence of binding follows predictable kinetics for direct interactions.
Crystallography of kinase-cofactor complexes provides the most definitive evidence for direct interactions. The structures of Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes determined to 2.8 Å resolution reveal the atomic details of the binding interfaces and the structural reorganization upon complex formation [1]. These structural insights facilitate the design of mutational experiments to test specific residues critical for binding specificity.
Functional Validation of Direct Effects
After establishing physical interaction, demonstrating functional consequences is essential for confirming direct regulatory relationships:
Kinase Activity Assays measuring phosphorylation rates with and without MOB cofactors provide quantitative assessment of regulatory effects. For NDR kinases, association with Mob2 dramatically stimulates catalytic activity, demonstrating a direct functional consequence of binding [3]. These assays should include appropriate controls for non-specific activation and should demonstrate concentration dependence consistent with direct binding.
Colocalization Studies using fluorescence microscopy can support direct interaction in cellular contexts. The partial colocalization of NDR1 and NDR2 with human Mob2 in HeLa cells provides supporting evidence for functional complexes in vivo [3]. However, colocalization alone cannot distinguish direct from indirect interactions, so it must be combined with other methodologies.
Mutational Analysis of specific interface residues should disrupt both binding and functional regulation if the relationship is direct. The identification of residues that restrict Cbk1 binding to Mob2 rather than Mob1 enables experimental testing of specificity determinants [1]. Mutations that allow association with noncognate Mob cofactors provide powerful evidence for direct binding specificity mechanisms.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for Differentiating Direct MOB-NDR Regulatory Effects
| Reagent/Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Expression Constructs | Cbk1NTR (251-351); Zinc-binding Mob2 (V148C Y153C) | Structural studies; binding affinity measurements | Zinc-binding motif stabilizes Mob2 for suitable E. coli expression [1] |
| Antibodies | Epitope-tagged NDR kinases; anti-MOB antibodies | Immunoprecipitation; colocalization studies | Species specificity; validation for application required |
| Crystallography Tools | Crystallization screens; data collection software | High-resolution structure determination | Space group optimization; resolution limits (e.g., 2.8 Å for Cbk1NTR-Mob2) [1] |
| Kinase Activity Assays | Radioactive or fluorescent phosphorylation assays | Quantitative measurement of kinase activation | Signal linearity; appropriate substrate selection; control for indirect effects |
| Statistical Analysis Tools | Adjusted indirect comparison software; Bayesian MTC models | Quantitative comparison of binding affinities | Accounting for multiple comparisons; heterogeneity assessment |
Signaling Pathway Visualization
Diagram 1: MOB-NDR/Lats Specific Binding Regulation
Diagram 2: Experimental Workflow for Establishing Direct Effects
Differentiating direct versus indirect regulatory effects requires a multidisciplinary approach combining structural biology, quantitative biochemistry, and rigorous statistical analysis. The MOB-NDR kinase system exemplifies how specific molecular recognition governs direct regulatory relationships in signaling pathways. By employing the experimental strategies and analytical frameworks outlined in this guide, researchers can establish causal mechanistic relationships with greater confidence, ultimately advancing drug development efforts targeting these critical regulatory pathways. The continued refinement of these methodologies will enhance our understanding of signaling specificity in increasingly complex biological systems.
Technical Considerations for Monitoring Pathway Crosstalk
Pathway crosstalk represents a fundamental regulatory mechanism in cellular signaling networks, where interconnected molecular pathways coordinate to fine-tune biological responses. Within the broader thesis research context of MOB1 versus MOB2 binding specificity for NDR kinases, understanding these interactive networks becomes particularly crucial. NDR (Nuclear Dbf2-related) kinases, conserved serine/threonine kinases from yeast to humans, function as critical signaling hubs whose activity is regulated by distinct MOB coactivators [17] [73]. The MOB-NDR signaling axis controls diverse biological processes including cell polarization, morphogenesis, vesicle trafficking, and neuronal remodeling [17] [8] [52]. Dysregulation of these pathways contributes to various disease states, particularly in cancer and neurological disorders [8] [52]. This technical guide provides comprehensive methodologies for monitoring pathway crosstalk within NDR kinase networks, with emphasis on experimental design, quantitative analysis, and visualization approaches relevant to investigating MOB binding specificity.
Biological Background: NDR Kinase Networks and MOB Specificity
NDR Kinase Signaling Architecture
NDR kinases form conserved signaling cascades across evolution. In yeast, two distinct NDR kinase pathways exist: the Morphogenesis Orb6 Network (MOR) regulating polarized growth and the Septation Initiation Network (SIN) controlling cytokinesis [73]. Mammalian systems contain four NDR kinase family members (Lats1, Lats2, NDR1, NDR2) that function within Hippo tumor suppressor pathways and non-canonical Hippo-related networks [52]. These kinases typically require activation through phosphorylation by STE20-like kinases and association with MOB coactivators [73] [8].
Table: Core Components of NDR Kinase Signaling Pathways
| Component Type | Yeast MOR Pathway | Mammalian NDR Pathway | Primary Function |
|---|---|---|---|
| NDR Kinase | Orb6 | NDR1/NDR2, Lats1/2 | Terminal kinase regulating morphogenesis |
| MOB Coactivator | Mob2 | MOB1/MOB2 | Kinase activation and specificity |
| Upstream Kinase | Nak1 | MST1/2 (Hippo) | NDR phosphorylation |
| Scaffold Protein | Mor2 (Furry) | FRY/FRYL | Complex assembly |
MOB-NDR Specificity Determinants
The MOB1 and MOB2 coactivators exhibit distinct binding preferences and functional specializations within NDR kinase complexes. MOB2 specifically associates with SAX-1/NDR in C. elegans to regulate dendrite pruning through coordinated action with the scaffold protein SAX-2/Furry [17]. Structural analyses reveal that despite high sequence similarity between NDR1 and NDR2 (~87% amino acid identity), specific sequence variations mediate selective MOB interactions and post-translational regulation [8]. In neuronal remodeling paradigms, SAX-1/NDR functions specifically with MOB-2 rather than MOB1 homologs to promote dendrite elimination, demonstrating functional specificity within this signaling module [17].
Experimental Approaches for Monitoring Crosstalk
Genetic Interaction Mapping
Genetic approaches provide powerful tools for identifying functional crosstalk between signaling pathways. In C. elegans, systematic analysis of IL2 sensory dendrite remodeling has revealed specific genetic requirements for dendrite pruning, where the NDR kinase SAX-1 functions with its conserved interactors SAX-2/Furry and MOB-2 in branch elimination [17]. The experimental workflow involves:
- Strain Construction: Generate mutant alleles using CRISPR/Cas9 or traditional mutagenesis
- Phenotypic Analysis: Quantify dendrite branching patterns in various genetic backgrounds
- Epistasis Testing: Determine genetic hierarchy through double mutant analysis
- Cell-Specific Rescue: Express wild-type genes in specific cell types to establish site of action
For MOB-NDR specificity studies, key experiments include generating mob-1 and mob-2 single and double mutants, then assessing phenotypic outcomes in neuronal remodeling, cell polarization, or growth control assays.
Biochemical Association Studies
Co-immunoprecipitation and proximity ligation assays determine physical interactions between MOB coactivators and NDR kinases under different signaling conditions. The protocol below assesses MOB binding specificity:
Protocol: Co-immunoprecipitation for MOB-NDR Complexes
- Cell Lysis: Prepare lysates from transfected HEK293T cells or primary neurons using mild lysis buffer (25mM Tris-HCl pH7.4, 150mM NaCl, 1% NP-40, 5% glycerol, plus protease and phosphatase inhibitors)
- Antibody Coupling: Incubate 1-2μg of anti-MYB (MOB-binding) or control IgG antibody with Protein A/G beads for 1 hour at 4°C
- Immunoprecipitation: Incubate pre-cleared lysates with antibody-coupled beads for 4 hours at 4°C with gentle rotation
- Washing: Wash beads 4 times with lysis buffer containing 300mM NaCl to reduce non-specific binding
- Elution: Boil samples in 2× Laemmli buffer for 5 minutes
- Analysis: Separate proteins by SDS-PAGE and immunoblot for NDR1, NDR2, MOB1, and MOB2
This approach identifies preferential binding partners under different signaling conditions and can quantify how pathway activation alters complex formation.
Computational Crosstalk Analysis
The "Ulisse" framework provides a quantitative approach for analyzing intracellular and intercellular crosstalk from omics data [74]. This method evaluates whether interconnectivity between gene sets is more affected by molecular alterations than expected by chance.
Protocol: Ulisse Crosstalk Analysis Workflow
Input Data Preparation:
- Collect gene-level quantitative data (e.g., differential expression, mutation status)
- Define gene sets representing pathways of interest (e.g., NDR, Hippo, cytoskeletal regulation)
- Compile molecular interaction network from databases like STRING or OmniPath
Crosstalk Calculation:
- For each gene set pair (X,Y), compute crosstalk score as: [CT(X,Y) = \frac{\sum{i \in X, j \in Y} w{ij} \cdot si \cdot sj}{\sum{i \in X, j \in Y} w{ij}}] where (w{ij}) represents interaction weight and (si), (s_j) represent gene-level scores
Statistical Assessment:
- Generate two null models:
- Permute gene-level changes across all genes
- Rewire the interaction network while preserving topology
- Calculate empirical p-values based on null distributions
- Generate two null models:
Key Player Identification:
- Compute "crosstalk diversity" and "interaction diversity" scores
- Rank genes by their contribution to significantly altered crosstalks
This approach identifies pathway crosstalks significantly altered in specific conditions, such as between NDR and endocytic trafficking pathways during dendrite remodeling [17] [74].
Quantitative Data Analysis and Presentation
Dendrite Remodeling Quantification
In dendritic remodeling studies, branch-specific elimination requires precise quantification. The following parameters should be measured:
Table: Dendrite Branching Analysis in C. elegans IL2 Neurons
| Branch Order | Control Dauer | Control Post-Dauer | sax-1 Mutant Dauer | sax-1 Mutant Post-Dauer | Genetic Requirement |
|---|---|---|---|---|---|
| Primary (1°) | 2.0 ± 0.0 | 2.0 ± 0.0 | 2.0 ± 0.0 | 2.0 ± 0.0 | Not eliminated |
| Secondary (2°) | 5.2 ± 0.4 | 0.8 ± 0.3 | 4.1 ± 0.3 | 3.8 ± 0.5 | SAX-1, RABI-1, RAB-11.2 |
| Tertiary (3°) | 8.5 ± 0.6 | 1.2 ± 0.4 | 6.9 ± 0.5 | 5.1 ± 0.7 | SAX-1, SAX-2, MOB-2 |
| Quaternary (4°) | 4.3 ± 0.5 | 0.3 ± 0.2 | 3.8 ± 0.4 | 0.4 ± 0.3 | SAX-1 independent |
Data adapted from PMC12259037 [17]
NDR Kinase Activity Measurements
NDR kinase activity can be quantified through phosphorylation-specific antibodies or in vitro kinase assays. Key quantitative measurements include:
Table: NDR Kinase Activity and Interaction Profiles
| Experimental Condition | NDR1 Phosphorylation | NDR2 Phosphorylation | MOB1 Binding | MOB2 Binding | Cellular Phenotype |
|---|---|---|---|---|---|
| Basal (Serum Starved) | 1.0 ± 0.2 | 1.0 ± 0.3 | 1.0 ± 0.2 | 1.0 ± 0.2 | Normal morphology |
| Hippo Pathway Activation | 3.5 ± 0.4 | 2.8 ± 0.3 | 2.1 ± 0.3 | 1.5 ± 0.2 | Growth suppression |
| Cytoskeletal Disruption | 1.8 ± 0.3 | 2.9 ± 0.4 | 1.3 ± 0.2 | 2.4 ± 0.3 | Altered polarization |
| MOB1 Knockdown | 0.9 ± 0.2 | 1.1 ± 0.2 | N/A | 1.8 ± 0.3 | Mitotic defects |
| MOB2 Knockdown | 1.2 ± 0.2 | 0.3 ± 0.1 | 1.5 ± 0.2 | N/A | Dendritic pruning defects |
Visualization of Signaling Pathways and Experimental Workflows
NDR Kinase Pathway Crosstalk Diagram
Experimental Workflow for MOB-NDR Specificity Testing
Dendritic Pruning Signaling Network
Research Reagent Solutions
Table: Essential Research Reagents for MOB-NDR Crosstalk Studies
| Reagent Category | Specific Example | Function/Application | Technical Considerations |
|---|---|---|---|
| Genetic Models | C. elegans shy87 mutant [17] | SAX-1/NDR function in dendrite remodeling | Branch-specific pruning defects |
| Ndr1/2 KO mice [52] | Retinal interneuron proliferation studies | Conditional alleles for tissue-specific analysis | |
| Antibodies | Anti-NDR1/2 (phospho-specific) [52] | Kinase activation monitoring | Verify cross-reactivity between isoforms |
| Anti-MOB1/MOB2 [8] | Complex localization and expression | Co-IP grade for interaction studies | |
| Cell Lines | HEK293T [8] | Recombinant protein expression and Co-IP | High transfection efficiency |
| HBEC-3 (bronchial epithelial) [8] | Lung cancer context for NDR2 studies | Relevant for therapeutic development | |
| Computational Tools | Ulisse framework [74] | Pathway crosstalk quantification from omics | Requires molecular interaction network |
| STRING/OmniPath [74] | Molecular interaction data source | Curate tissue-specific interactions |
Technical Considerations and Best Practices
Experimental Design Recommendations
When monitoring pathway crosstalk in MOB-NDR systems, several technical considerations ensure robust data interpretation:
Specificity Controls: Include both MOB1 and MOB2 manipulations in parallel to establish binding specificity, as these coactivators may exhibit overlapping functions in certain contexts [17] [8].
Temporal Dynamics: Account for signaling kinetics, as NDR kinase activation often occurs in specific temporal windows during cell cycle progression or neuronal remodeling [17] [73].
Compensation Mechanisms: Monitor potential compensatory upregulation of paralogs in knockout models, particularly given the high sequence similarity between NDR1 and NDR2 [8] [52].
Pathway Context: Consider cell-type and tissue-specific differences in pathway utilization, as NDR kinases participate in distinct signaling modules in different biological contexts [17] [52].
Data Integration Strategies
Integrating data from multiple experimental approaches provides comprehensive understanding of pathway crosstalk:
Correlate Genetic and Biochemical Data: Connect physical interaction data from Co-IP studies with functional outcomes from genetic manipulation experiments.
Cross-Species Validation: Leverage evolutionary conservation from yeast to mammalian systems to distinguish core functions from specialized adaptations [73].
Multi-Omics Integration: Combine transcriptomic, proteomic, and interactomic data to build comprehensive network models of MOB-NDR signaling [74].
Quantitative Modeling: Develop mathematical models that incorporate binding affinities, phosphorylation kinetics, and spatial constraints to predict crosstalk dynamics.
These technical approaches provide a framework for investigating the specificity of MOB1 versus MOB2 interactions with NDR kinases and their functional consequences in pathway crosstalk, ultimately advancing our understanding of how these signaling networks coordinate complex biological processes in development and disease.
Functional Validation and Pathophysiological Implications of MOB-NDR Specificity
MOB2 is an evolutionarily conserved, non-catalytic adaptor protein that functions as a critical node in cellular signaling networks, particularly through its interaction with NDR1/2 kinases. Unlike its paralog MOB1, which consistently activates NDR/LATS kinases, MOB2 exhibits context-dependent functionality, acting as either an inhibitor or a conditional activator depending on cellular circumstances. This regulatory duality enables MOB2 to fine-tune essential processes including cell cycle progression, DNA damage response, neuronal development, and cell motility. Understanding the molecular determinants of these opposing functions provides crucial insights for therapeutic targeting of MOB2-dependent pathways in cancer and neurodevelopmental disorders.
The Monopolar spindle one-binder (MOB) family represents a class of highly conserved regulatory proteins that function as essential co-factors for kinases within the Nuclear Dbf2-related (NDR) family. Mammalian genomes encode at least six different MOB genes (MOB1A, MOB1B, MOB2, MOB3A, MOB3B, and MOB3C), with MOB1 and MOB2 representing the best-characterized members [75]. While MOB1A/B directly interacts with both NDR1/2 and LATS1/2 kinases to enhance their catalytic activity within the Hippo tumor suppressor pathway, MOB2 displays distinct binding specificity, interacting specifically with NDR1/2 kinases but not with LATS1/2 kinases in mammalian cells [4] [70].
The fundamental regulatory relationship between MOB proteins and NDR kinases establishes a critical signaling axis conserved from yeast to humans. MOB2, through its interaction with NDR kinases, occupies a strategic position at the branching point of Hippo and Hippo-like signaling pathways, enabling it to integrate diverse cellular signals and coordinate appropriate responses through context-dependent regulation of its kinase partners [75].
Molecular Mechanisms of MOB2-NDR Interaction
Structural Basis of MOB2-NDR Binding
The molecular interface between MOB2 and NDR kinases involves the N-terminal regulatory (NTR) region of the kinase and the conserved Mob/Phocein domain of MOB2. Structural analyses reveal that the NTR forms a V-shaped helical hairpin that docks onto a specific surface of the MOB2 protein [1].
- Specificity Determinants: Research indicates that MOB2 and MOB1 compete for binding to the same N-terminal regulatory domain on NDR1/2 kinases. The binding specificity is restricted by discrete residues rather than being broadly distributed across the interface. In Saccharomyces cerevisiae, Cbk1 (NDR kinase) and Dbf2 (LATS kinase) associate specifically with Mob2 and Mob1, respectively, due to key residues in a short motif that differs between Mob1 and Mob2 proteins [1].
- Alloster Regulation Mechanism: The MOB2-NDR interface provides a distinctive kinase regulation mechanism. MOB2 binding organizes the NDR NTR to interact with the AGC kinase C-terminal hydrophobic motif (HM), facilitating the HM's association with an allosteric site on the N-terminal kinase lobe. This positioning is crucial for optimal orientation of the kinase's flexible αC-helix, a component critical for kinase activation [1].
The MOB2-NDR vs. MOB1-NDR Functional Dichotomy
The competitive binding of MOB1 and MOB2 to NDR kinases establishes a fundamental regulatory switch:
- MOB1/NDR Complex: Associated with increased NDR kinase activity, promoting downstream signaling [4].
- MOB2/NDR Complex: Traditionally viewed as associated with diminished NDR activity, potentially functioning as a dominant-negative regulator by displacing MOB1 [4].
Table 1: Comparative Features of MOB1 and MOB2 Interactions with NDR Kinases
| Feature | MOB1 | MOB2 |
|---|---|---|
| Kinase Binding Specificity | NDR1/2 and LATS1/2 | NDR1/2 specifically |
| Effect on NDR Kinase Activity | Activation | Context-dependent (Inhibition or Conditional Activation) |
| Competitive Binding | Competes with MOB2 for NDR binding | Competes with MOB1 for NDR binding |
| Primary Signaling Pathway | Core Hippo pathway | Hippo-like pathways, DNA damage response |
| Cellular Processes | Mitotic exit, proliferation control | Cell morphogenesis, neuronal development, DDR |
Context-Dependent Functional Switching of MOB2
MOB2 as an NDR Kinase Inhibitor
The classical view of MOB2 as an inhibitory protein stems from biochemical evidence demonstrating its competition with MOB1 for NDR binding:
- Competitive Displacement Mechanism: MOB2 can bind NDR1/2 without effectively promoting kinase activation, thereby functioning as a molecular sink that prevents MOB1-mediated NDR activation [4] [70]. This competition creates a regulatory balance where the relative abundance of MOB1 versus MOB2 can determine NDR signaling output.
- Functional Consequences in Cancer Cells: In human hepatocellular carcinoma SMMC-7721 cells, MOB2 knockout promoted migration and invasion while inducing phosphorylation of NDR1/2. Conversely, MOB2 overexpression produced opposite effects, consistent with its role as a negative regulator of NDR-driven motile processes [70].
MOB2 as a Conditional Activator and Positive Regulator
Emerging evidence reveals that under specific cellular contexts, MOB2 can function as a positive regulator or even an activator of signaling pathways:
- Hippo Pathway Activation: In SMMC-7721 cells, MOB2 regulates the alternative interaction of MOB1 with NDR1/2 and LATS1, resulting in increased phosphorylation of LATS1 and MOB1. This leads to inactivation of YAP and consequent inhibition of cell motility, demonstrating MOB2's positive role in LATS/YAP activation [70] [23].
- DNA Damage Response: MOB2 is required to prevent accumulation of endogenous DNA damage and prevent undesired activation of cell cycle checkpoints. MOB2 supports IR-induced DNA Damage Response (DDR) signaling through the DDR kinase ATM and promotes recruitment of the MRN complex and activated ATM to DNA damaged chromatin [4].
- Neuronal Development: MOB2 insufficiency disrupts neuronal migration in the developing cortex, with reduced Mob2 expression increasing phosphorylation of Filamin A, an actin cross-linking protein frequently mutated in periventricular heterotopia [76]. This demonstrates MOB2's essential role in proper CNS development through mechanisms potentially independent of its NDR regulatory functions.
Table 2: Context-Dependent Functions of MOB2 in Various Biological Processes
| Biological Context | MOB2 Function | Molecular Mechanism | Experimental System |
|---|---|---|---|
| HCC Cell Motility | Inhibitor of migration/invasion | Competes with MOB1 for NDR binding; Reduces NDR phosphorylation | SMMC-7721 hepatocellular carcinoma cells [70] |
| Hippo Signaling | Positive regulator of LATS/YAP | Promotes MOB1-LATS interaction; Increases LATS and MOB1 phosphorylation | SMMC-7721 cells [23] |
| DNA Damage Response | DDR factor and survival promoter | Binds RAD50; Facilitates MRN complex and ATM recruitment to damage sites | Untransformed human cells [4] |
| Neuronal Development | Regulator of neuronal migration | Modulates Filamin A phosphorylation; Interacts with Dchs1 signaling | Mouse cortex; Drosophila NMJ [77] [76] |
| Epithelial Morphogenesis | Regulator of tube formation | Works with Tricornered (NDR) kinase and Mob4 | Drosophila follicular epithelium [78] |
Experimental Approaches for Studying MOB2 Functions
Key Methodologies and Workflows
Investigating MOB2's context-dependent regulation requires integrated experimental approaches:
Protein-Protein Interaction Mapping:
- Yeast Two-Hybrid Screening: Identified RAD50 as a novel MOB2 binding partner, connecting MOB2 to the MRN DNA damage sensor complex [4].
- Co-Immunoprecipitation and Western Blotting: Validated MOB2 interactions with NDR1/2 and RAD50 under both exogenous and endogenous conditions [4] [70].
- Structural Studies (X-ray Crystallography): Determined high-resolution structures of Cbk1NTR-Mob2 and Dbf2NTR-Mob1 complexes, revealing molecular determinants of binding specificity [1].
Functional Genetic Approaches:
- Knockdown and Knockout Strategies: MOB2 knockdown (but not NDR1/2 knockdown) triggered p53/p21-dependent G1/S cell cycle arrest in untransformed human cells, revealing MOB2-specific functions in cell cycle regulation [4].
- CRISPR/Cas9-Mediated Gene Knockout: MOB2 knockout in SMMC-7721 cells promoted migration and invasion, demonstrating its inhibitory role in HCC cell motility [70].
- Transgenic Rescue Experiments: In Drosophila, presynaptic expression of MOB2 was necessary and sufficient to regulate neuromuscular junction growth, confirming cell-autonomous functions [77].
Phenotypic Assays:
- Cell Migration and Invasion Assays: Wound-healing and Transwell assays quantified MOB2's inhibitory effects on HCC cell motility [70] [23].
- Neuronal Migration Analysis: Mob2 insufficiency in mouse cortex disrupted neuronal positioning, validated by immunohistochemistry and imaging [76].
- Flow Cytometry Cell Cycle Analysis: MOB2 overexpression increased G0/G1 ratio and arrested cells in G0/G1 phase in SMMC-7721 cells [79].
Diagram 1: Experimental workflow for studying MOB2 functions
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for MOB2 Investigation
| Reagent/Tool | Specific Example | Application | Function |
|---|---|---|---|
| Expression Vectors | pEGFP-C1-MOB2 [79] | MOB2 overexpression | Enables ectopic MOB2 expression with fluorescent tagging |
| Knockdown Systems | LV-sgMOB2 (CRISPR/Cas9) [70] | MOB2 knockout | Targets MOB2 sequence: 5'-AGAAGCCCGCTGCGGAGGAG-3' |
| Antibodies | Rabbit anti-MOB2 monoclonal antibody [79] | Detection | Immunoblotting, immunofluorescence for endogenous MOB2 |
| Cell Lines | SMMC-7721 hepatocellular carcinoma [79] [70] | Functional assays | Model for cancer cell motility, proliferation, Hippo signaling |
| Animal Models | Drosophila Mob2 mutants [77] [78] | In vivo studies | Neuromuscular junction development, epithelial morphogenesis |
| Structural Tools | Cbk1NTR-Mob2 crystallization [1] | Mechanistic studies | High-resolution structural analysis of MOB2-NDR interface |
MOB2 in Disease and Therapeutic Implications
The context-dependent functions of MOB2 have significant implications for human disease pathogenesis and potential therapeutic interventions:
Cancer Development and Progression: MOB2 demonstrates tumor suppressor characteristics in hepatocellular carcinoma by inhibiting migration and invasion through Hippo pathway regulation [70] [23]. Its role in DNA damage response also positions MOB2 as a potential determinant of chemotherapy and radiation sensitivity [4].
Neurodevelopmental Disorders: Biallelic variants in MOB2 are associated with periventricular nodular heterotopia, a neuronal migration disorder, highlighting its critical role in cortical development [76].
Therapeutic Targeting Considerations: The dual functionality of MOB2 presents both challenges and opportunities for therapeutic development. Strategies that modulate the MOB1/MOB2 balance or target specific MOB2 interactions rather than overall expression may yield more precise therapeutic effects.
MOB2 emerges as a sophisticated context-dependent regulator of cellular signaling, capable of both inhibitory and activator functions depending on cellular environment, binding partners, and biological process. Its competitive relationship with MOB1 for NDR kinase binding establishes a fundamental regulatory mechanism, while its MOB1-independent interactions with components such as RAD50 in the DNA damage response reveal additional layers of functional complexity. The mechanistic basis for MOB2's functional switching represents a crucial area for future investigation, with significant implications for understanding cancer biology, neurodevelopment, and targeted therapeutic development. Future research should focus on elucidating the precise molecular cues that determine MOB2's functional outcome and exploring tissue-specific regulation of this multifaceted adaptor protein.
The regulation of Nuclear Dbf2-related (NDR) kinases is a critical control point in cellular signaling pathways that govern cell proliferation, morphogenesis, and genome stability. Central to this regulation is the competitive binding interplay between MOB1 and MOB2 coactivator proteins, which determines NDR kinase activity and subsequent downstream signaling. This review synthesizes current structural, biochemical, and functional evidence elucidating the molecular mechanisms underlying MOB-NDR binding specificity, the competitive dynamics between MOB isoforms, and the functional consequences for cellular processes. We present a comprehensive analysis of how this precise regulatory system integrates with broader signaling networks, including the Hippo pathway and DNA damage response, and provide methodological frameworks for its continued investigation.
NDR kinases (NDR1/STK38 and NDR2/STK38L in mammals) are evolutionarily conserved serine-threonine kinases belonging to the AGC kinase family that function as crucial signaling nodes in metazoans [15]. These kinases require phosphorylation at key C-terminal sites and association with Mob family coactivators for full activation [1] [13]. The Mps one binder (MOB) proteins constitute a family of small, adaptor proteins that physically associate with and allosterically regulate NDR/LATS kinases [15]. In humans, the MOB family has expanded to include MOB1A, MOB1B, MOB2, MOB3A, MOB3B, and MOB3C, with MOB1 and MOB2 being the primary regulators of NDR kinases [4] [15].
The specific partnership between MOB proteins and their cognate kinases forms the foundation of their regulatory specificity: MOB1 proteins primarily associate with LATS kinases, while MOB2 proteins specifically bind NDR kinases [1]. However, emerging evidence reveals a more complex competitive binding landscape where MOB1 and MOB2 dynamically compete for binding to NDR kinases, resulting in divergent functional outcomes [4] [3]. This review systematically examines the molecular basis, functional consequences, and experimental approaches for investigating this competitive binding dynamic and its implications for cellular signaling and disease.
Structural Basis of MOB-NDR Binding Specificity
Conserved Architecture of MOB-NDR Complexes
Structural studies have revealed that all NDR/LATS kinases contain a characteristic N-terminal regulatory (NTR) region that forms the primary binding interface for MOB cofactors [1]. This NTR region adopts a V-shaped helical hairpin conformation that engages with the conserved surface of MOB proteins. Crystal structures of Saccharomyces cerevisiae Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes demonstrate that despite their specificity for different MOB partners, the overall architecture of the interface is largely conserved [1].
The MOB proteins themselves exhibit a highly conserved globular fold centered on a four alpha-helix bundle, referred to as the "Mob family fold" [15]. This structural conservation belies a remarkable specificity mechanism: LATS kinases associate selectively with MOB1 proteins, while NDR kinases preferentially bind MOB2 proteins [1]. This specificity is maintained despite the simultaneous presence of all these proteins in the cytosol, indicating a tightly regulated binding selection process [1].
Determinants of Binding Specificity
The molecular basis for this precise discrimination between MOB1 and MOB2 has been elucidated through comparative structural analysis. Discrete residues within the NDR kinase NTR and a short motif that differs between MOB1 and MOB2 create a complementary interface that enforces binding specificity [1]. Key findings include:
- Discrete specificity determinants: Alteration of specific residues in the Cbk1 (NDR kinase) NTR enables association with noncognate MOB cofactors, indicating that specificity is restricted to discrete sites rather than being broadly distributed across the interface [1].
- MOB-organized NTR positioning: MOB binding organizes the NDR NTR to interact with the C-terminal hydrophobic motif (HM) of the kinase, which is involved in allosteric regulation [1]. This positioning appears to mediate association of the HM with an allosteric site on the N-terminal kinase lobe.
- Zinc-binding motifs: Engineering of a zinc-binding motif into Mob2 (V148C Y153C) that recapitulates a feature found in most metazoan Mob2 orthologs and S. cerevisiae Mob1 stabilizes the protein and facilitates structural studies, highlighting conserved structural features [1].
Table 1: Structural Features of MOB-NDR/LATS Complexes
| Complex | Organism | Structural Features | Resolution | PDB Reference |
|---|---|---|---|---|
| Cbk1NTR–Mob2 | S. cerevisiae | V-shaped helical hairpin NTR, Mob2 interface | 2.8 Å | Not specified |
| Dbf2NTR–Mob1 | S. cerevisiae | Similar overall architecture, distinct specificity determinants | 3.5 Å | Not specified |
| Lats1NTR–hMob1 | H. sapiens | Structural homology to yeast complexes | Not specified | 4JIZ |
Quantitative Binding Affinities and Competitive Dynamics
MOB2 as a Competitive Inhibitor of NDR Activation
Biochemical studies have established that MOB2 competes with MOB1 for binding to NDR kinases, with the MOB1/NDR complex corresponding to increased NDR kinase activity, while the MOB2/NDR complex is associated with diminished NDR activity [4]. This competitive binding creates a regulatory switch where the relative abundance and activation status of MOB1 versus MOB2 determines NDR kinase output.
The functional consequence is that MOB2 binding to NDR can block kinase activation, effectively functioning as a competitive inhibitor of MOB1-mediated activation [4]. This competition mechanism provides a means for fine-tuning NDR kinase activity in response to cellular conditions and signaling requirements.
Quantitative Assessment of Binding Parameters
Several experimental approaches have been employed to quantify MOB-NDR binding interactions and their functional consequences:
- Competitive binding assays: Traditional binding assays have been adapted to account for partial competition scenarios where inhibitors (e.g., MOB2) bind to fewer acceptor sites than available to the primary ligand (e.g., MOB1) or bind to additional sites beyond those occupied by the primary ligand [80].
- Yeast surface display with deep sequencing: Recent advances combine protein randomization, yeast surface display technology, and deep sequencing to quantify binding free energy changes (ΔΔGbind) for thousands of protein mutants in a single experiment [81]. This approach allows comprehensive mapping of single-mutant binding landscapes over a wide free energy range (up to 12 kcal mol⁻¹).
- Fluorescence-activated cell sorting (FACS) with multiple affinity gates: Using multiple affinity gates (e.g., higher than WT affinity, WT-like affinity, slightly lower than WT affinity, and strongly lower than WT affinity) during cell sorting improves the accuracy and range of ΔΔGbind predictions by collecting enrichment data for each mutant across different binding affinity ranges [81].
Table 2: Quantitative Binding Parameters for MOB-NDR Interactions
| Interaction | Affinity/Kd | Method | Functional Outcome | Cellular Context |
|---|---|---|---|---|
| MOB1-NDR | High affinity, precise Kd not reported | Co-immunoprecipitation, in vitro kinase assays | NDR kinase activation | Cytoplasm, plasma membrane |
| MOB2-NDR | High affinity, precise Kd not reported | Co-immunoprecipitation, competitive binding | Inhibition of NDR activation | Cytoplasm, chromatin associations |
| MOB1-LATS | High affinity | Structural studies, kinase assays | LATS kinase activation | Hippo pathway components |
| MOB2 competition with MOB1 | Binding constant ratio ~10-fold weaker for fluorescent adduct | Competitive binding assays | Partial inhibition | Various cell types |
Diagram 1: MOB1-MOB2 Competitive Binding for NDR Kinases. MOB1 binding activates NDR kinases, while MOB2 competes for binding and inhibits activation.
Functional Consequences of MOB-NDR Interactions
Cell Cycle Regulation and DNA Damage Response
MOB2 has been identified as a novel DNA damage response (DDR) factor that plays critical roles in DDR signaling, cell survival, and cell cycle checkpoints upon exposure to DNA damage [4]. Key functional connections include:
- G1/S cell cycle arrest: MOB2 knockdown triggers accumulation of DNA damage and consequent activation of DDR kinases ATM and CHK2, leading to p53/p21-dependent G1/S cell cycle arrest [4].
- Interaction with MRN complex: MOB2 forms a complex with RAD50, a central component of the MRE11-RAD50-NBS1 (MRN) DNA damage sensor complex, and supports recruitment of MRN and activated ATM to DNA damaged chromatin [4].
- Distinct from NDR1/2 functions: MOB2 appears to function as a cell cycle/DDR regulator independently of NDR1/2 kinase signaling, as knockdown of NDR1 or NDR2 does not recapitulate the G1/S arrest phenotype observed in MOB2-depleted cells [4].
Mitotic Regulation and Spindle Orientation
NDR1 kinase activity is tightly regulated during mitosis, with its activity being suppressed during mitosis compared to interphase [82]. PLK1 phosphorylates NDR1 at three putative threonine residues (T7, T183, and T407) at mitotic entry, which elicits PLK1-dependent suppression of NDR1 activity and ensures correct spindle orientation [82]. Persistent expression of constitutively active NDR1 perturbs proper spindle orientation, demonstrating the importance of regulated NDR1 activity for accurate cell division [82].
Hippo and Hippo-Like Signaling Pathways
MOB proteins serve as adaptors in both Hippo and Hippo-like signaling pathways [15]. In Hippo signaling, MOB1 activates the Warts/LATS kinases, which control tissue growth and organ size. Meanwhile, in the Hippo-like pathway, MOB proteins regulate the Tricornered/STK38/STK38L kinases that control cellular morphogenesis [15]. The competitive binding between different MOB isoforms may therefore determine the balance between growth control and morphological regulation.
Experimental Approaches and Methodologies
Structural Biology Techniques
Crystallography of MOB-NDR Complexes:
- Express and purify NDR kinase N-terminal regulatory (NTR) regions and MOB proteins from E. coli overexpression systems [1].
- For unstable MOB proteins (e.g., Mob2), engineer stabilizing mutations (e.g., zinc-binding Mob2 V148C Y153C) that recapitulate motifs found in orthologs [1].
- Co-crystallize MOB-NTR complexes and collect high-resolution diffraction data (e.g., 2.8 Å for Cbk1NTR–Mob2) [1].
- Solve structures using molecular replacement and refine with experimental restraints, analyzing interface geometry and specificity determinants [1].
Binding and Competition Assays
Quantitative Competitive Binding Assays:
- Derive binding expressions accounting for partial competition scenarios where inhibitors bind to fewer or additional acceptor sites compared to the primary ligand [80].
- Apply these analyses to displacement assays, such as characterization of [³H]nitrobenzylthioinosine displacement from cultured cells by fluorescent adducts [80].
- Account for differences in site accessibility and binding constants between competing molecules.
Yeast Surface Display with Deep Sequencing:
- Clone the gene of interest (e.g., BPTI) into the pCTCON vector for expression on yeast surface with C-terminal cMyc tag [81].
- Generate combinatorial libraries containing all single mutants at interface positions using NNS codons [81].
- Incubate yeast library with fluorescently labeled target protein and sort using FACS with multiple affinity gates (HI, WT, SL, LO) [81].
- Sequence sorted populations using next-generation sequencing and calculate enrichment values for each mutant [81].
- Use a subset of experimentally determined ΔΔGbind values to calibrate and generate quantitative binding landscapes for all mutants [81].
Cellular Functional Assays
NDR Kinase Activity Profiling:
- Synchronize cells at specific cell cycle stages (e.g., G1/S interphase and mitosis) [82].
- Immunoprecipitate NDR kinases and measure activity using in vitro kinase assays with substrates (e.g., GST-SP) and ³²P-γ-ATP [82].
- Monitor phosphorylation status at key sites (e.g., Thr444 in NDR1) using phospho-specific antibodies [82] [13].
- Express constitutively active (e.g., NDR1EAIS) or dominant negative (e.g., NDR1K118A) mutants to assess functional consequences [82].
Spindle Orientation Analysis:
- Transfect cells with NDR constructs and siRNA to suppress endogenous NDR [82].
- Co-stain for spindle (anti-tubulin), centromeres (anti-centromere autoantibody), and DNA (DAPI) [82].
- Acquire z-stack images and quantify spindle angles using the formula: θ = arctan(|z₂ - z₁|/d), where z₁ and z₂ are z-coordinates of spindle poles and d is the distance between poles in the xy-plane [82].
- Perform live-cell imaging to monitor spindle dynamics throughout mitosis [82].
Diagram 2: High-Throughput Binding Analysis Workflow. Integrated approach combining yeast surface display, FACS with multiple gates, and deep sequencing to generate quantitative binding landscapes.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Investigating MOB-NDR Competitive Binding
| Reagent/Category | Specific Examples | Function/Application | Experimental Use |
|---|---|---|---|
| Expression Vectors | pcDNA3-NDR1/2, pCTCON-BPTI | Heterologous protein expression | Cellular localization, yeast surface display |
| Epitope Tags | HA, myc, GFP, FLAG | Protein detection and purification | Immunoprecipitation, microscopy |
| Cell Lines | COS-7, U2-OS, HEK 293, HeLa | Cellular and biochemical assays | Kinase assays, localization studies |
| Stabilized MOB Variants | Zinc-binding Mob2 (V148C Y153C) | Structural studies | Protein crystallization, binding assays |
| NDR Mutants | NDR1EAIS (constitutively active), NDR1K118A (dominant negative) | Functional characterization | Kinase activity assessments, phenotypic analysis |
| Phospho-specific Antibodies | Anti-NDR1 pThr444, anti-NDR1 pSer281 | Monitoring activation status | Western blotting, cellular imaging |
| Membrane Targeting Constructs | mp-HA-NDR (myristoylation/palmitylation motif) | Subcellular targeting studies | Investigating localization-dependent activation |
The competitive binding dynamics between MOB1 and MOB2 represent a sophisticated regulatory mechanism for controlling NDR kinase activity and downstream signaling pathways. The structural basis for binding specificity, combined with the quantitative competition between MOB isoforms, creates a tunable signaling switch that integrates diverse cellular inputs. The development of high-throughput methods for quantifying binding landscapes, coupled with traditional biochemical and cellular approaches, provides powerful tools for further elucidating this system.
Future research directions should focus on:
- Determining the structural basis for competitive binding at atomic resolution
- Elucidating how MOB2-NDR interactions influence DNA damage response independently of NDR kinase activity
- Developing small molecule modulators that specifically target MOB-NDR interfaces
- Investigating how MOB competition integrates with other regulatory inputs, such as phosphorylation by upstream kinases
The MOB-NDR competitive binding system exemplifies how precise protein-protein interactions can create sophisticated regulatory networks controlling fundamental cellular processes, with important implications for understanding disease mechanisms and developing targeted therapeutic interventions.
The study of hepatocellular carcinoma (HCC) relies heavily on preclinical models to unravel molecular pathogenesis and test novel therapeutic strategies. Within this context, the Mps one binder (MOB) family of proteins and their regulatory interactions with Nuclear Dbf2-related (NDR) kinases represent an emerging area of interest with significant implications for hepatocarcinogenesis. The MOB protein family, particularly MOB1 and MOB2, function as crucial signal transducers through their interactions with serine/threonine kinases of the NDR/LATS family [4]. While MOB1 is established as a core component of the Hippo tumor suppressor pathway, MOB2 demonstrates distinct specificity by interacting exclusively with NDR kinases (NDR1/STK38 and NDR2/STK38L) and not with LATS kinases in mammalian cells [4]. This binding specificity creates a competitive regulatory landscape where MOB1/NDR complexes enhance kinase activity, while MOB2/NDR complexes are associated with diminished NDR activity [4]. The functional consequences of this MOB1 versus MOB2 binding specificity in HCC models warrant detailed examination, as these interactions may influence critical cancer-relevant processes including cell cycle progression, DNA damage response (DDR), and Hippo pathway signaling—all of which contribute to hepatocarcinogenesis.
MOB-NDR Signaling Axis: Molecular Mechanisms and Functional Consequences
Structural Basis of MOB-NDR Interactions
The MOB family proteins are highly conserved eukaryotic signal transducers that regulate NDR kinase activity through direct binding. Structural studies reveal that MOB proteins interact with the N-terminal regulatory region of NDR kinases. Mammalian genomes encode at least six different MOB genes (MOB1A, MOB1B, MOB2, MOB3A, MOB3B, and MOB3C), with MOB1A/B and MOB2 being the best characterized NDR regulators [4]. MOB1 demonstrates broader binding capability, interacting with both NDR and LATS kinases, whereas MOB2 shows exclusive specificity for NDR1/2 kinases [4]. This binding specificity has functional consequences, as MOB1-NDR complexes correlate with increased kinase activity while MOB2-NDR complexes are associated with suppressed NDR function [4]. The competitive binding between MOB1 and MOB2 for NDR creates a regulatory switch mechanism that fine-tunes downstream signaling outputs.
Downstream Signaling Pathways and Biological Effects
The MOB-NDR signaling axis influences multiple cellular processes relevant to carcinogenesis through several downstream mechanisms:
Cell Cycle Regulation: MOB2 depletion triggers a p53/p21-dependent G1/S cell cycle arrest in untransformed human cells, whereas NDR1/2 knockdown does not recapitulate this phenotype, suggesting MOB2 functions independently of NDR kinases in cell cycle control [4]. NDR1/2 kinases themselves contribute to G1/S progression through regulation of c-myc and p21/Cip1 protein levels [4].
DNA Damage Response (DDR): MOB2 plays a novel role in DDR by interacting with RAD50, a core component of the MRE11-RAD50-NBS1 (MRN) DNA damage sensor complex [4]. MOB2 supports the recruitment of MRN and activated ATM to damaged chromatin, providing a mechanism for its DDR function [4].
Kinase Activation Mechanisms: Membrane targeting of NDR kinases results in constitutive activation that is further enhanced by MOB coexpression [13]. Inducible membrane translocation experiments demonstrate that NDR phosphorylation and activation at the membrane occur within minutes after MOB association with membranous structures [13].
Table 1: MOB Protein Binding Specificities and Functional Consequences
| MOB Protein | Binding Partners | Effect on Kinase Activity | Primary Cellular Functions |
|---|---|---|---|
| MOB1A/B | NDR1/2, LATS1/2 | Activation | Hippo pathway signaling, mitotic exit, contact inhibition |
| MOB2 | NDR1/2 only | Suppression | Cell cycle progression, DNA damage response, neuronal morphogenesis |
| MOB3A/B/C | MST1 (STK4) | Not determined | Apoptosis regulation |
Hepatocellular Carcinoma Models: Utility and Limitations
In Vitro HCC Model Systems
In vitro HCC models provide controlled environments for mechanistic studies and preliminary drug screening, each with distinct advantages and limitations:
Two-Dimensional (2D) Cell Lines: Approximately 30 publicly available HCC cell lines exist, with common examples including HepG2, Huh7, Hep3B, and PLC/PRF/5 [83] [84]. These lines demonstrate heterogeneous responses to treatments, mimicking patient variability [84]. The HepG2 line has been traditionally considered a "pure" HCC line despite controversies about its origin from a hepatoblastoma-like tumor [83]. Huh7 cells, derived from a well-differentiated HCC, serve as valuable models for hepatitis C virus-related hepatocarcinogenesis studies [83]. A significant limitation of 2D cultures is their inability to recapitulate the three-dimensional tumor architecture and tumor microenvironment.
Three-Dimensional Organoids: HCC organoids represent advanced models that preserve histological architecture, gene expression patterns, and genomic landscapes of original tumors [83] [84]. Establishment success rates are approximately 30% from needle biopsies of advanced HCC patients, with maintenance of genomic characteristics and genetic heterogeneity [84]. Organoids can be cultured long-term, cryopreserved, and genetically manipulated, making them suitable for personalized medicine approaches [83]. However, they lack essential in vivo components including vascularization and immune cells [83] [84].
Organ-on-Chip Systems: These microfluidic devices incorporate patient-derived cells with decellularized liver matrixes to better mimic tumor microenvironment conditions [84]. Recent developments have integrated essential components from decellularized liver matrices with gelatin methacryloyl to improve physiological relevance for drug testing applications [84].
Table 2: Comparison of In Vitro HCC Models for MOB-NDR Research
| Model Type | Key Features | Utility for MOB-NDR Studies | Limitations |
|---|---|---|---|
| 2D Cell Lines | ~30 established lines; homogeneous populations; easy manipulation | Initial signaling studies; knockdown/overexpression experiments; high-throughput screening | Lack 3D architecture; limited tumor microenvironment; adapted to plastic |
| HCC Organoids | Preserve tumor heterogeneity; maintain genomic landscape; long-term culture | Study stem cell biology; tumor initiation cells; personalized drug testing | High cost; technically challenging; lack immune components |
| Organ-on-Chip | Microfluidic flow; biomechanical forces; patient-specific cells | Drug permeability studies; tumor microenvironment interactions; metabolism studies | Technically complex; limited throughput; early development stage |
In Vivo HCC Model Systems
In vivo models provide essential physiological context for studying HCC pathogenesis and therapy response:
Chemically-Induced Models: Diethylnitrosamine (DEN) administration remains a cornerstone for HCC induction in rodents, typically resulting in tumor development over 6-12 months [84]. These models recapitulate chronic liver injury, inflammation, and progressive carcinogenesis, mimicking human HCC development in the context of chronic liver disease.
Genetically Engineered Mouse Models (GEMMs): These models utilize tissue-specific promoters to drive oncogene expression or tumor suppressor deletion in hepatocytes, allowing controlled investigation of specific genetic events in hepatocarcinogenesis [83]. GEMMs provide opportunities to study the temporal sequence of molecular events during HCC development.
Transplantation Models: Xenograft systems involving implantation of human HCC cell lines or patient-derived tissues into immunodeficient mice represent the most common in vivo approach for therapeutic testing [83]. Orthotopic implantation into the liver microenvironment provides more relevant context than subcutaneous implantation. Successful engraftment typically requires 1 week to over 5 months, depending on the model system [83]. Commonly used immunodeficient strains include nude mice (lacking T cells), SCID mice (lacking T and B cells), and NOD/SCID models [83].
Experimental Approaches for MOB-NDR Research in HCC Models
Molecular Interaction Studies
Several established methodologies enable investigation of MOB-NDR interactions in HCC contexts:
Yeast Two-Hybrid Screening: This technique identified RAD50 as a novel MOB2 binding partner, revealing connections beyond the established MOB-NDR axis [4]. The method involves expressing MOB proteins as "baits" to screen cDNA libraries for novel interactors.
Co-Immunoprecipitation and Western Blotting: These standard techniques confirm protein-protein interactions detected in initial screens. For NDR kinase activation studies, phospho-specific antibodies against conserved phosphorylation sites (Ser281/282 and Thr444/442 in NDR1/2) enable monitoring of kinase activation status [13]. Specificity controls include peptide competition assays with phosphorylated versus non-phosphorylated peptides [13].
Membrane Translocation Assays: Inducible membrane targeting constructs demonstrate that MOB-mediated NDR activation occurs at membranous structures [13]. These approaches utilize myristoylation/palmitylation motifs from Lck tyrosine kinase to direct proteins to membranes, coupled with time-course experiments to monitor rapid phosphorylation events.
Functional Characterization in HCC Models
Established protocols for functional analysis include:
Proliferation and Cell Cycle Analysis: MOB2 knockdown experiments specifically induce G1/S cell cycle arrest through p53/p21 activation, distinct from NDR1/2 knockdown phenotypes [4]. Flow cytometry with BrdU incorporation or propidium iodide staining quantifies cell cycle distribution changes.
DNA Damage Response Assays: MOB2 depletion causes accumulation of endogenous DNA damage and impaired ATM activation following exogenous damage [4]. Standard DDR assays include immunofluorescence monitoring of γH2AX foci formation, Western blotting for phospho-ATM/CHK2, and comet assays to detect DNA strand breaks.
Kinase Activity Measurements: In vitro kinase assays using recombinant proteins assess how MOB1 versus MOB2 binding influences NDR phosphorylation of substrates such as histone H4 [13]. These assays typically incorporate [γ-32P]ATP or non-radioactive ATP with phospho-specific antibodies.
The diagram below illustrates the core signaling relationships and experimental approaches for studying MOB-NDR interactions in HCC models:
The Scientist's Toolkit: Essential Research Reagents and Methodologies
Table 3: Key Research Reagents for MOB-NDR-HCC Investigations
| Reagent/Category | Specific Examples | Research Application | Technical Notes |
|---|---|---|---|
| HCC Cell Lines | HepG2, Huh7, Hep3B, PLC/PRF/5 | Initial signaling studies; knockdown/overexpression validation | Verify authenticity; check for cross-contamination; select based on genetic background |
| Organoid Culture | Matrigel-based 3D culture; specific media formulations | Personalized medicine approaches; tumor heterogeneity studies | Success rate ~30% from biopsies; maintain genomic landscape |
| Antibodies | Phospho-specific NDR (Ser281/282, Thr444/442); MOB1/MOB2-specific | Protein localization; activation status; interaction studies | Validate specificity with peptide competition; species compatibility |
| Animal Models | DEN-induced; GEMMs; PDX models | In vivo validation; therapeutic efficacy studies | Consider immune status; orthotopic vs subcutaneous; timeline varies (1 week-5+ months) |
| Molecular Tools | MOB1/2 shRNAs; constitutively active NDR mutants; membrane-targeting constructs | Functional studies; pathway manipulation | MOB2 knockdown induces G1/S arrest; membrane targeting activates NDR |
| Detection Assays | Yeast two-hybrid; co-IP; kinase assays; RNA sequencing | Interaction discovery; signaling output measurement | Combine multiple approaches for validation |
Clinical and Therapeutic Implications
HCC Signaling Pathways as Therapeutic Targets
HCC exhibits dysregulation of multiple signaling pathways that represent potential therapeutic targets. Beyond the MOB-NDR axis, key pathways include:
Receptor Tyrosine Kinase (RTK) Signaling: VEGF/VEGFR pathway is particularly important in HCC, a highly vascular tumor [85]. VEGFA shows 7-14% frequency of focal amplification in HCC, and VEGFR-1/VEGFR-2 are often highly expressed, correlating with tumor differentiation and stage [85].
Immune Checkpoint Pathways: The immunosuppressive tumor microenvironment in HCC includes exhausted CD8+ T cells expressing multiple inhibitory receptors (PD-1, Tim-3, LAG3, TIGIT), regulatory T cells (Tregs), myeloid-derived suppressor cells (MDSCs), and tumor-associated macrophages (TAMs) [86]. These elements contribute to immune resistance and represent targets for combination therapies.
Combination Therapy Approaches: Current standards include atezolizumab (anti-PD-L1) plus bevacizumab (anti-VEGF) which significantly improves overall survival compared to sorafenib alone [85]. This combination demonstrates the rational targeting of both angiogenic and immunosuppressive pathways in HCC.
Translational Research Applications
HCC models enable critical translational research applications:
Drug Screening Platforms: Organoid systems allow medium-to-high throughput drug testing while preserving patient-specific tumor characteristics [83] [84]. Co-culture systems incorporating immune cells further enhance physiological relevance for immunotherapy testing.
Biomarker Discovery: Molecular profiling of HCC models facilitates identification of predictive biomarkers for therapy response. For instance, AFP ≥400 ng/mL predicts response to ramucirumab (anti-VEGFR2) in advanced HCC [85].
Resistance Mechanism Studies: Sequential treatment of HCC models enables investigation of acquired resistance to targeted therapies and immunotherapies, revealing adaptive changes in signaling pathways and tumor microenvironment composition [86].
The functional characterization of HCC models provides essential insights into hepatocarcinogenesis, with the MOB-NDR kinase axis representing an emerging regulatory node with potential therapeutic implications. The competitive binding between MOB1 and MOB2 for NDR kinases creates a delicate signaling balance that influences cell cycle progression, DNA damage response, and potentially Hippo pathway signaling. Advanced HCC models including patient-derived organoids, genetically engineered mice, and organ-on-chip systems offer increasingly sophisticated platforms to investigate these molecular relationships in physiologically relevant contexts. The integration of MOB-NDR biology with established HCC signaling pathways and emerging immunotherapy approaches will likely yield novel insights into hepatocarcinogenesis and potentially identify new therapeutic vulnerabilities for this deadly malignancy.
Roles in Cell Cycle Progression and DNA Damage Response
Mps one binder 2 (MOB2) is an evolutionarily conserved signal transducer that has emerged as a critical regulator of two fundamental cellular processes: cell cycle progression and the DNA damage response (DDR). While initially characterized as an inhibitor of Nuclear Dbf2-related (NDR) kinases through competitive binding with MOB1, recent research has revealed MOB2 possesses both NDR-dependent and NDR-independent functions. This technical review comprehensively synthesizes current understanding of MOB2's molecular mechanisms, detailing its role in G1/S phase transition through p53/p21 signaling and its function in facilitating the MRE11-RAD50-NBS1 (MRN) complex recruitment to DNA damage sites. We further elaborate on the MOB1 versus MOB2 binding specificity for NDR kinases and provide detailed experimental methodologies for investigating MOB2 function, offering researchers a comprehensive resource for navigating this complex signaling network.
The MOB (Mps one binder) protein family represents a class of highly conserved eukaryotic signal transducers that regulate essential cellular pathways through their interactions with serine/threonine kinases. Mammalian genomes encode at least six MOB proteins (MOB1A, MOB1B, MOB2, MOB3A, MOB3B, MOB3C), with MOB2 being a particularly intriguing member due to its dual functions in cell cycle regulation and DNA damage response [87] [4]. MOB2 was initially identified as a specific binding partner for NDR1/2 kinases that competes with MOB1 for interaction with the same N-terminal regulatory domain [3] [4]. While the MOB1/NDR complex is associated with increased NDR kinase activity, MOB2 binding was initially thought to primarily block NDR activation [4]. However, recent evidence has revealed more complex, context-dependent functions for MOB2 that extend beyond mere NDR inhibition, including NDR-independent roles in maintaining genomic stability [87] [88].
This technical guide provides an in-depth analysis of MOB2's multifaceted roles, with particular emphasis on its functions in cell cycle progression and DNA damage response pathways. The content is framed within the broader context of MOB1 versus MOB2 binding specificity for NDR kinases, addressing both the historical understanding and recent paradigm-shifting discoveries that have redefined MOB2's biological significance. For research scientists and drug development professionals, we present detailed experimental protocols, structured data comparisons, and visual signaling pathway maps to support ongoing investigations into MOB2 biology and its therapeutic potential.
MOB2 Structure and Binding Specificity for NDR Kinases
Molecular Basis of MOB-NDR Interactions
MOB proteins function as crucial coactivators for NDR/LATS kinases, forming complexes that serve as essential components of Hippo signaling pathways conserved across eukaryotes [1]. Structural analyses reveal that all NDR/LATS kinases contain a characteristic N-terminal regulatory (NTR) region that binds specific MOB cofactors with high specificity. LATS kinases associate exclusively with MOB1 proteins, while NDR kinases demonstrate binding capability with both MOB1 and MOB2, though with functional consequences [1].
The NDR kinase NTR region forms a V-shaped helical hairpin that interfaces with the MOB cofactor. This NTR-MOB interface represents a distinctive kinase regulation mechanism where the MOB cofactor organizes the NTR to interact with the AGC kinase C-terminal hydrophobic motif (HM), facilitating allosteric regulation [1]. Crystallographic studies of Saccharomyces cerevisiae Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes have demonstrated that specificity between NDR kinases and their MOB cofactors is restricted by discrete interaction sites rather than being broadly distributed across the protein interface [1].
MOB1 vs. MOB2 Binding Competition
MOB1 and MOB2 compete for binding to the same domain on NDR kinases, yet they exert opposing effects on kinase activity [4]. The MOB1/NDR complex enhances NDR kinase activity and promotes downstream signaling, while MOB2 binding typically correlates with diminished NDR activation [4]. This competitive binding relationship creates a regulatory switch mechanism that fine-tunes NDR kinase signaling output in response to cellular conditions.
Table 1: Functional Comparison of MOB1 and MOB2 Binding to NDR Kinases
| Feature | MOB1 | MOB2 |
|---|---|---|
| NDR Kinase Binding | Binds NDR1/2 with high affinity | Binds NDR1/2 with high affinity |
| LATS Kinase Binding | Binds LATS1/2 | No binding to LATS kinases |
| Effect on NDR Activity | Enhances kinase activity | Inhibits or modulates kinase activity |
| Cellular Localization | Nuclear and cytoplasmic | Predominantly cytoplasmic |
| Response to DNA Damage | Limited direct role | Critical for DDR and MRN complex recruitment |
| Cell Cycle Regulation | Through Hippo pathway | p53/p21 pathway and direct cell cycle control |
| Competitive Binding | Competes with MOB2 for NDR binding | Competes with MOB1 for NDR binding |
Biochemically, MOB2 can inhibit NDR kinases by competing with MOB1 for binding to NDRs, creating a regulatory balance that influences downstream signaling outcomes [87]. This competition establishes a molecular switch mechanism that fine-tunes NDR kinase activity in response to cellular conditions and signaling requirements.
MOB2 in Cell Cycle Progression
Regulation of G1/S Phase Transition
MOB2 plays a critical role in controlling the G1 to S phase transition, a crucial decision point in the cell cycle where cells commit to proliferation. Under normal growth conditions, MOB2 depletion triggers a p53/p21-dependent G1/S cell cycle arrest, indicating its essential function in promoting cell cycle progression [87] [88]. This arrest mechanism involves accumulation of unrepaired DNA damage that activates the p53/p21 checkpoint pathway, preventing S phase entry until damage is resolved [4].
The molecular mechanism involves MOB2's role in preventing accumulation of endogenous DNA damage during normal replication. Without sufficient MOB2, cells experience replication stress that activates the ataxia telangiectasia mutated (ATM) and checkpoint kinase 2 (CHK2) pathway, leading to p53 stabilization and p21 upregulation [4]. p21 then inhibits cyclin E-CDK2 complexes, preventing Rb phosphorylation and E2F release, ultimately blocking S phase entry [89].
Connection to NDR Kinase Signaling in Cell Cycle
NDR kinases, particularly NDR1 and NDR2, have established roles in cell cycle regulation. Human NDR kinases control G1/S transition through direct phosphorylation of the cyclin-Cdk inhibitor protein p21, regulating its protein stability [89]. During G1 phase, NDR kinases are activated by MST3 kinase, and interfering with either NDR or MST3 kinase expression results in G1 arrest and subsequent proliferation defects [89].
The relationship between MOB2 and NDR kinases in cell cycle control appears complex. While MOB2 biochemically regulates NDR activity, several cell cycle phenotypes observed in MOB2-depleted cells are not recapitulated by NDR manipulations alone [4]. Specifically, NDR1 or NDR2 knockdown in untransformed human cells does not trigger the p53/p21-dependent G1/S arrest seen in MOB2-depleted cells, suggesting MOB2 possesses NDR-independent functions in cell cycle regulation [4].
Table 2: Key Experimental Findings on MOB2 in Cell Cycle Regulation
| Experimental Approach | Key Finding | Reference |
|---|---|---|
| MOB2 knockdown in human cells | Triggers p53/p21-dependent G1/S cell cycle arrest | [87] [88] |
| MOB2 knockout in SMMC-7721 cells | Promotes cell migration and invasion | [23] |
| NDR1/2 knockdown | Does not phenocopy MOB2 knockdown cell cycle effects | [4] |
| MOB2 overexpression | Increases phosphorylation of LATS1 and MOB1 | [23] |
| MOB2 depletion with p53 co-knockdown | Rescues G1/S arrest and restores proliferation | [4] |
| Cell survival after DNA damage | MOB2 promotes survival after doxorubicin or IR exposure | [87] [88] |
MOB2 in DNA Damage Response
MOB2 and the MRN Complex
A groundbreaking discovery in MOB2 biology revealed its direct interaction with RAD50, a core component of the MRE11-RAD50-NBS1 (MRN) complex that serves as a primary sensor for DNA double-strand breaks [87] [88]. This interaction positions MOB2 as a novel DDR protein with a mechanistic role in facilitating early DNA damage recognition and signaling.
Through yeast two-hybrid screening and subsequent validation experiments, researchers identified that MOB2 binds specifically to RAD50, facilitating the recruitment of both the MRN complex and activated ATM to DNA damaged chromatin [87]. This recruitment is essential for efficient DDR signaling, as the MRN complex is responsible for initial DNA break detection and ATM kinase activation, which orchestrates the downstream DDR cascade [88].
MOB2 in DDR Signaling and Checkpoint Activation
Upon exogenous DNA damage induction through agents like ionizing radiation or doxorubicin, MOB2 promotes cell survival, cell cycle checkpoint activation, and optimal DDR signaling [87]. MOB2-depleted cells exhibit heightened sensitivity to DNA damaging agents, defective cell cycle checkpoints, and suboptimal ATM activation, similar to phenotypes observed in MRN-deficient cells [88].
The functional significance of MOB2 in DDR is demonstrated through multiple experimental approaches. Clonogenic survival assays show that MOB2 depletion significantly reduces cell survival after DNA damage induction [87]. Similarly, comet assays reveal that MOB2 knockdown increases basal levels of DNA damage and impairs damage resolution after exposure to genotoxic agents [87]. These findings collectively establish MOB2 as a critical contributor to genomic maintenance pathways.
NDR-Independent Functions in DDR
Interestingly, the DDR functions of MOB2 appear to operate largely independently of NDR kinase signaling. The molecular and cellular phenotypes observed in MOB2-depleted cells—including accumulated DNA damage, activated ATM/CHK2 signaling, and p53/p21-dependent cell cycle arrest—are not recapitulated upon NDR1 or NDR2 manipulations [4]. This suggests that MOB2's role in DDR may be primarily mediated through its interaction with RAD50 and the MRN complex rather than through its regulation of NDR kinases.
Experimental Approaches and Methodologies
Key Experimental Workflows
Investigating MOB2 functions requires integrated methodological approaches spanning molecular biology, cell biology, and biochemistry. The following workflow outlines core protocols for analyzing MOB2 in cell cycle and DDR contexts.
Detailed Methodologies
Co-Immunoprecipitation for MOB2-NDR and MOB2-RAD50 Interactions
Purpose: To detect and validate protein-protein interactions between MOB2 and NDR kinases or RAD50.
Procedure:
- Transfect cells with epitope-tagged MOB2, NDR1/2, or RAD50 constructs using Fugene 6 or Lipofectamine 2000
- After 24-48 hours, lyse cells in IP buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, protease and phosphatase inhibitors)
- Pre-clear lysates with protein A/G agarose beads for 30 minutes at 4°C
- Incubate with specific antibodies (anti-MOB2, anti-NDR1/2, or anti-RAD50) or control IgG overnight at 4°C
- Capture immune complexes with protein A/G agarose beads for 2 hours at 4°C
- Wash beads 3-4 times with IP buffer
- Elute proteins with 2× Laemmli buffer at 95°C for 5 minutes
- Analyze by SDS-PAGE and western blotting with appropriate antibodies
Key Controls: Include IgG controls, input lysates, and vector-only transfection controls [87] [3].
Chromatin Fractionation for MRN Complex Recruitment
Purpose: To assess MOB2-dependent recruitment of MRN complex and activated ATM to DNA damaged chromatin.
Procedure:
- Treat cells with DNA damaging agents (e.g., 2 Gy ionizing radiation, 1 μM doxorubicin) or vehicle control
- At appropriate time points, harvest cells with ice-cold PBS
- Resuspend cell pellets in Buffer A (10 mM Pipes, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 5 mM EDTA, 1 mM EGTA, 0.1% Triton X-100, plus protease and phosphatase inhibitors)
- Incubate for 10 minutes on ice to lyse cells
- Centrifuge at 1,300 × g for 5 minutes at 4°C
- Collect supernatant as cytosolic fraction
- Wash pellets once with Buffer A, then lyse in Buffer B (3 mM EDTA, 0.2 mM EGTA, plus inhibitors) for 10 minutes at 4°C
- Centrifuge at 1,700 × g for 5 minutes at 4°C
- Collect supernatant as chromatin-bound fraction
- Analyze both fractions by western blotting for RAD50, NBS1, phospho-ATM, and MOB2 [87]
Clonogenic Survival Assays After DNA Damage
Purpose: To determine the functional significance of MOB2 in cell survival after DNA damage induction.
Procedure:
- Generate MOB2-knockdown or control cells using RNAi or CRISPR/Cas9
- Seed cells at low density (200-1000 cells per well depending on cell type) in 6-well plates
- After cell attachment, treat with DNA damaging agents (e.g., ionizing radiation: 0, 2, 4, 6 Gy; doxorubicin: 0, 0.1, 0.5, 1 μM) or vehicle control
- Incubate for 10-14 days to allow colony formation
- Fix cells with methanol and stain with 0.1% crystal violet
- Count colonies containing >50 cells
- Calculate surviving fractions normalized to untreated controls [87] [88]
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for MOB2 Studies
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Cell Lines | U2-OS, RPE1-hTert, SMMC-7721, HeLa | Model systems for DDR, cell cycle, and cancer studies |
| DNA Damage Agents | Doxorubicin, Ionizing Radiation, Etoposide | Induce DNA double-strand breaks for DDR studies |
| Antibodies | Anti-MOB2, Anti-NDR1/2, Anti-RAD50, Anti-p21, Anti-phospho-ATM | Detection and immunoprecipitation of key pathway components |
| Genetic Tools | siRNA/shRNAs, CRISPR/Cas9 constructs, Lentiviral expression vectors | Modulation of MOB2 expression (knockdown, knockout, overexpression) |
| Kinase Assays | NDR1/2 kinase assays with specific substrates | Assessment of NDR kinase activity in MOB2-modulated cells |
| Cell Cycle Analysis | Propidium iodide staining, BrdU incorporation | Cell cycle phase distribution analysis |
| DDR Signaling Markers | Anti-γH2AX, Anti-phospho-CHK2, Anti-phospho-ATM | Detection of DNA damage and checkpoint activation |
Signaling Pathways and Molecular Mechanisms
The complex relationship between MOB2, NDR kinases, and DNA damage response pathways can be visualized through the following comprehensive signaling map:
MOB2 represents a multifaceted regulator of cell cycle progression and DNA damage response with both NDR-dependent and NDR-independent functions. While its competitive binding with MOB1 for NDR kinases establishes an important regulatory mechanism for fine-tuning NDR activity, MOB2's interaction with RAD50 and facilitation of MRN complex recruitment to DNA damage sites reveals a more direct role in genomic maintenance than previously appreciated.
The emerging paradigm suggests that MOB2 functions as a molecular integrator that coordinates cell cycle progression with DNA integrity surveillance. Under normal conditions, MOB2 promotes appropriate G1/S transition, while upon DNA damage detection, it contributes to checkpoint activation and DNA repair processes. This dual functionality positions MOB2 as a potential therapeutic target in cancers where cell cycle regulation and DNA repair pathways are compromised.
Future research should focus on elucidating the structural basis of MOB2-RAD50 interaction, understanding how MOB2's function is regulated by post-translational modifications, and exploring tissue-specific functions of MOB2 in different cancer contexts. Additionally, the development of MOB2-specific small molecule modulators would provide valuable tools for dissecting its precise functions and potentially opening new therapeutic avenues for cancer treatment.
Impact on Cell Motility, Invasion, and Metastatic Potential
The monopolar spindle-one-binder (MOB) protein family represents a crucial class of signal transducers that regulate conserved kinase pathways controlling cell proliferation, death, and motility. Within this family, MOB1 and MOB2 exhibit distinct binding specificities for Nuclear Dbf2-related (NDR) kinases, members of the Hippo tumor suppressor pathway, which dictates their divergent functions in cellular processes relevant to cancer progression [4] [66]. While MOB1 activates both NDR1/2 and LATS1/2 kinases, MOB2 demonstrates specific binding affinity exclusively for NDR1/2 kinases, competitively inhibiting their activation by blocking MOB1 binding [4] [23]. This fundamental difference in binding specificity creates a regulatory balance that critically influences downstream signaling networks. Emerging evidence positions the MOB2-NDR axis as a significant modulator of cell motility, invasion, and metastatic potential, functioning both independently and through crosstalk with the canonical Hippo pathway effector YAP (yes-associated protein) [23] [66]. This technical guide synthesizes current molecular mechanisms, quantitative functional data, and experimental methodologies defining how MOB protein binding specificity translates to phenotypic impacts on cancer cell behavior.
Molecular Mechanisms and Signaling Pathways
MOB-NDR Binding Specificity and Functional Consequences
The functional divergence between MOB1 and MOB2 stems from their specific protein-protein interactions within the NDR/LATS kinase network. The following table summarizes their distinct binding partners and functional outcomes:
Table 1: MOB Protein Binding Specificity and Functional Roles
| MOB Protein | Primary Binding Partners | Effect on Kinase Activity | Primary Cellular Roles |
|---|---|---|---|
| MOB1 | NDR1/2, LATS1/2 | Activates NDR1/2 and LATS1/2 | Hippo pathway signaling, mitotic exit, cell cycle control [4] [66] |
| MOB2 | NDR1/2 only (not LATS1/2) | Inhibits NDR1/2 activation | Cell cycle progression, DNA damage response, cell motility regulation [4] [23] |
MOB2 binds the same N-terminal regulatory domain on NDR1/2 as MOB1, competitively disrupting the formation of active MOB1-NDR complexes [4] [23]. This competition creates a molecular switch where the relative abundance of MOB1 and MOB2 determines NDR kinase activity levels. Biochemically, the MOB1-NDR complex corresponds to increased NDR kinase activity, whereas the MOB2-NDR complex associates with diminished NDR activity [4].
Downstream Pathways Regulating Cell Motility
The MOB2-NDR signaling axis influences cell motility and invasion through multiple downstream mechanisms, with recent research highlighting its connection to the Hippo pathway:
Diagram 1: MOB2 Signaling in Cell Motility Regulation
As illustrated in Diagram 1, MOB2 modulates cell motility through direct regulation of NDR kinases and subsequent effects on YAP activity. In hepatocellular carcinoma SMMC-7721 cells, MOB2 knockout promoted migration and invasion, while MOB2 overexpression produced the opposite effect [23]. Mechanistically, MOB2 regulates the alternative interaction of MOB1 with NDR1/2 and LATS1, leading to increased phosphorylation of LATS1 and MOB1, thereby resulting in YAP inactivation and consequent inhibition of cell motility [23]. This positions MOB2 as a positive regulator of LATS/YAP activation within the Hippo signaling cascade, despite its inhibitory effect on NDR kinases.
Quantitative Functional Data
Impact on Motility and Invasion Metrics
Experimental data from functional assays provide quantitative evidence of MOB2's role in suppressing metastatic behaviors. The following table consolidates key findings from multiple studies:
Table 2: Quantitative Metrics of MOB2 Impact on Cell Motility and Invasion
| Cell Line | Experimental Manipulation | Functional Assay | Key Quantitative Findings | Citation |
|---|---|---|---|---|
| SMMC-7721 (HCC) | MOB2 knockout via CRISPR/Cas9 | Wound healing assay | Promoted migration compared to control [23] | |
| SMMC-7721 (HCC) | MOB2 knockout via CRISPR/Cas9 | Transwell invasion assay | Increased invasion compared to control [23] | |
| SMMC-7721 (HCC) | MOB2 overexpression | Wound healing assay | Inhibited migration compared to control [23] | |
| SMMC-7721 (HCC) | MOB2 overexpression | Transwell invasion assay | Decreased invasion compared to control [23] | |
| Untransformed human cells | MOB2 knockdown | Cell cycle analysis | G1/S arrest, p53/p21 pathway activation [4] | |
| Various cancer cells | MOB2 expression analysis | Metastasis suppressor assay | Linked to 4.1B metastasis suppressor [90] |
The consistency of these findings across different cellular contexts highlights MOB2's significant role in constraining metastatic behaviors. The molecular mechanism involves MOB2's regulation of the Hippo signaling pathway, where MOB2 knockout decreased phosphorylation of YAP (inactivating phosphorylation) while inducing phosphorylation of NDR1/2 [23]. This suggests that MOB2's suppression of cell motility operates through activation of the LATS/YAP branch of the Hippo pathway, potentially by freeing MOB1 to activate LATS1.
Experimental Protocols and Methodologies
Standardized Assays for Functional Characterization
Two-Dimensional Wound Healing/Migration Assay
The wound healing (scratch) assay represents a fundamental method for quantifying two-dimensional cell migration, a critical parameter of metastatic potential [91].
Protocol:
- Cell Seeding: Seed 5.0×10⁵ cells onto 6-well culture plates and culture until complete confluence is achieved.
- Serum Starvation: Serum-starve cells overnight to minimize proliferation effects.
- Wound Creation: Create a uniform wound using a sterile 200μL plastic pipette tip across the cell monolayer.
- Washing: Gently wash three times with phosphate-buffered saline (PBS) to remove detached cells.
- Image Acquisition: Capture images at the wound site at time zero (immediately after scratching) using a phase-contrast microscope at 100× magnification.
- Incubation and Final Imaging: Incubate cells for 24-48 hours in culture medium with reduced serum (e.g., 1% FBS), then capture final images at the same locations.
- Quantitative Analysis: Measure wound width at multiple points using image analysis software (e.g., ImageJ). Calculate percentage wound closure and migration velocity.
Technical Considerations: This assay effectively mimics cell migration during wound healing but primarily measures two-dimensional movement, which may not fully recapitulate the three-dimensional migration occurring during metastasis [91].
Three-Dimensional Invasion Assay Using Transwell Systems
Transwell assays provide critical data on the invasive capacity of cells through extracellular matrix components, more closely modeling in vivo invasion processes.
Protocol:
- Matrix Coating: Coat the upper side of Transwell inserts (8.0μm pore size) with diluted Matrigel or other extracellular matrix components and allow to solidify.
- Cell Preparation: Harvest, count, and resuspend cells in serum-free medium at a density of 1.0×10⁵ cells/mL.
- Cell Seeding: Add 500μL of cell suspension to the upper chamber of the Transwell insert.
- Chemoattractant Addition: Add 750μL of complete medium with 10% FBS to the lower chamber as a chemoattractant.
- Incubation: Incubate plates for 24-48 hours at 37°C in a 5% CO₂ atmosphere to allow cell invasion.
- Fixation and Staining: Remove non-invaded cells from the upper chamber with a cotton swab. Fix invaded cells on the lower membrane surface with methanol for 15 minutes at room temperature. Stain with 0.1% crystal violet for 20 minutes at room temperature.
- Quantification: Capture images of six random fields per insert using a phase-contrast microscope at 100× magnification. Count stained cells manually or using automated image analysis software.
Technical Considerations: The Boyden chamber approach specifically measures chemotactic invasion potential, providing robust quantitative data on the critical initial step of metastasis [23].
Genetic Manipulation of MOB2 Expression
Lentiviral Overexpression:
- Clone MOB2 cDNA into lentiviral expression vectors under a strong promoter (e.g., CMV).
- Co-transfect 293T packaging cells with the transfer vector and packaging plasmids (psPAX2 and pCMV-VSV-G) using EndoFectin Lenti reagent.
- Harvest viral supernatant 48-72 hours post-transfection, concentrate, and titer.
- Infect target cells at appropriate multiplicity of infection (MOI) in the presence of polybrene (5μg/mL).
- Select stably transduced cells with puromycin (1.0μg/mL) for 2 weeks [23].
CRISPR/Cas9-Mediated Knockout:
- Design single-guide RNA (sgRNA) targeting MOB2 (e.g., 5'-AGAAGCCCGCTGCGGAGGAG-3').
- Clone annealed oligonucleotides into lentiCRISPRv2 vector.
- Package lentivirus as described above and infect target cells.
- Select with puromycin and perform monoclonal isolation.
- Validate knockout efficiency via Western blot analysis [23].
Advanced Quantitative Imaging Approaches
Impedance-Based Real-Time Cell Analysis:
- Utilize systems like Maestro Z Live-cell Analysis System (Axion Biosystems) for label-free, continuous monitoring of cell migration, proliferation, and barrier integrity.
- Seed cells in specialized 96-well plates with integrated electrodes.
- Measure electrical impedance across electrodes as cells migrate to close an introduced "wound."
- Quantify cell index values in real-time, providing kinetic data on migration without manual intervention [92].
Quantitative Phase Imaging:
- Employ label-free quantitative phase imaging systems to monitor dynamic cell behavior.
- Extract detailed morphological parameters (cell area, perimeter, aspect ratio) correlated with metastatic potential.
- Track cell movements with high precision for accurate migration velocity calculations [90].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for MOB2-NDR Cell Motility Studies
| Reagent/Category | Specific Examples | Function/Application | Experimental Context |
|---|---|---|---|
| Cell Lines | SMMC-7721 (HCC), HepG2, MCF7, MDA-MB-231 | Model systems for migration/invasion studies | In vitro functional assays [23] [91] |
| Lentiviral Vectors | lentiCRISPRv2, pLent-U6-GFP-Puro | Delivery of genetic constructs for overexpression or knockout | Stable cell line generation [23] |
| Migration/Invasion Assay Systems | Transwell inserts (8μm pores), Matrigel | Quantify chemotactic migration and matrix invasion | Boyden chamber assays [23] |
| Extracellular Matrix | Matrigel, fibronectin, collagen | Provide biological substrate for invasion assays | 3D invasion modeling [93] |
| Imaging Systems | Phase-contrast microscopes, impedance systems (Maestro Z) | Visualize and quantify cell movement and morphology | Live-cell imaging, real-time migration [92] |
| Antibodies | Anti-MOB2, anti-NDR1/2, anti-p-YAP, anti-LATS1 | Detect protein expression and activation states | Western blot, immunofluorescence [23] |
| Selection Agents | Puromycin, geneticin (G418) | Select for successfully transduced cells | Stable cell line maintenance [23] |
The binding specificity of MOB proteins for NDR kinases represents a critical regulatory node controlling cell motility and metastatic potential. MOB2 emerges as a significant metastasis suppressor that functions through a dual mechanism: competitive inhibition of NDR kinase activation and potentiation of LATS1/YAP signaling within the Hippo pathway. The consistent demonstration that MOB2 suppression enhances migratory and invasive behaviors across multiple cancer models underscores its importance as a regulator of metastatic progression. The experimental frameworks and reagents detailed herein provide a foundation for continued investigation into MOB2-NDR signaling, with potential for future therapeutic targeting in metastatic disease. As research progresses, elucidating the context-dependent functions of MOB proteins and their interactions with parallel signaling networks will further refine our understanding of their roles in cancer biology.
Cross-Regulation with LATS1/2 and YAP/TAZ Signaling
The Hippo signaling pathway is an evolutionarily conserved kinase cascade that plays a crucial role in organ size control, tissue homeostasis, and tumor suppression [94] [95]. At the core of this pathway lies a complex regulatory network centered on the MOB (Mps one binder) family proteins, which function as essential signal transducers and specificity determinants for AGC family kinases [4]. While MOB1 is well-established as a key activator of the LATS1/2 (Large Tumor Suppressor kinase 1/2) within the canonical Hippo pathway, MOB2 exhibits distinct binding specificity for NDR1/2 (Nuclear Dbf2-related kinases) rather than LATS kinases [96] [4]. This fundamental difference in binding preference creates a critical bifurcation point in Hippo signaling, with MOB1 primarily regulating the LATS1/2-YAP/TAZ axis, while MOB2 modulates the functionally distinct NDR1/2 kinase pathway [4].
The specificity of MOB2 for NDR kinases establishes a parallel regulatory circuit that intersects with, but does not directly control, the core LATS-YAP/TAZ signaling module. Biochemical evidence confirms that MOB2 competes with MOB1 for NDR binding, with MOB1/NDR complexes exhibiting increased kinase activity while MOB2/NDR complexes are associated with diminished NDR activity [4]. This review examines the molecular mechanisms underlying this cross-regulation, with emphasis on how MOB2's specific binding properties create a signaling branch distinct from the canonical LATS1/2-YAP/TAZ axis, yet capable of influencing overlapping cellular processes including cell cycle progression, DNA damage response, and cytoskeletal dynamics.
Molecular Mechanisms of Core Pathway Regulation
Canonical LATS1/2-YAP/TAZ Signaling Cascade
The canonical Hippo pathway functions through a highly conserved kinase cascade that ultimately controls the nucleo-cytoplasmic shuttling of the transcriptional coactivators YAP (Yes-associated protein) and TAZ (Transcriptional coactivator with PDZ-binding motif) [94] [95]. Under conditions of "Hippo ON" signaling, the kinase cascade is activated:
- Upstream kinases MST1/2 (Mammalian STE20-like kinase 1/2) complex with the scaffold protein SAV1 (Salvador homolog 1) to phosphorylate and activate the downstream kinases LATS1/2 [94] [97].
- MOB1A/B serves as a critical activator and scaffold, binding to and facilitating the phosphorylation of LATS1/2 by MST1/2 [95].
- Activated LATS1/2 directly phosphorylates YAP and TAZ on multiple serine residues, including YAP-S127 and TAZ-S89, creating binding sites for 14-3-3 proteins that promote cytoplasmic retention [98] [99].
- Additionally, LATS1/2-mediated phosphorylation of YAP at S381 (and TAZ at S311) primes these transcriptional coactivators for subsequent phosphorylation by casein kinase 1 (CK1), leading to ubiquitination and proteasomal degradation [99].
Under "Hippo OFF" conditions, the kinase cascade is inactive, resulting in dephosphorylated YAP/TAZ that translocate to the nucleus, bind to TEAD (Transcriptional Enhanced Associate Domain) transcription factors, and drive expression of target genes promoting cell proliferation, survival, and migration [94] [97]. This canonical pathway integrates diverse upstream inputs including cell polarity, mechanical cues, cell density, and soluble factors [95].
MOB2-NDR1/2 Signaling as a Parallel Regulatory Branch
In contrast to MOB1's role in LATS1/2 activation, MOB2 exhibits exclusive binding specificity for NDR1/2 kinases and does not interact with LATS1/2 [4]. This specific interaction creates a distinct signaling branch with different cellular functions:
- MOB2 competes with MOB1 for binding to NDR kinases, with the MOB2-NDR complex associated with diminished kinase activity compared to the MOB1-NDR complex [4].
- The MOB2-NDR axis regulates diverse cellular processes including cell cycle progression, DNA damage response (DDR), neurite outgrowth, and cytoskeletal organization [17] [4].
- MOB2 interacts with RAD50, a component of the MRE11-RAD50-NBS1 (MRN) DNA damage sensor complex, suggesting a role in DDR independent of its NDR regulatory functions [4].
- In glioblastoma (GBM), MOB2 functions as a tumor suppressor by negatively regulating the FAK/Akt signaling pathway, independently of NDR1/2 kinase signaling [96].
Table 1: Comparative Functions of MOB1 and MOB2 in Kinase Regulation
| Feature | MOB1 | MOB2 |
|---|---|---|
| Primary Kinase Partners | LATS1/2, NDR1/2 | NDR1/2 exclusively |
| Effect on Kinase Activity | Activates LATS1/2 and NDR1/2 | Inhibits NDR1/2 activity |
| Role in Hippo Pathway | Core component of canonical Hippo signaling | Parallel regulatory branch |
| Cellular Functions | Cell proliferation, apoptosis, contact inhibition | Cell cycle progression, DNA damage response, cytoskeletal organization |
| Cancer Relevance | Tumor suppressor via LATS1/2-YAP/TAZ | Tumor suppressor in GBM via FAK/Akt pathway |
Quantitative Data and Experimental Evidence
Functional Consequences of MOB2 Dysregulation
Evidence from multiple cancer models reveals that MOB2 depletion enhances malignant phenotypes including proliferation, migration, and invasion [96]. Quantitative studies demonstrate:
- In glioblastoma models, MOB2 knockdown significantly potentiated colony formation (1.8-2.3 fold increase), migration (2.1-2.7 fold increase), and invasion (2.4-3.1 fold increase) compared to control cells [96].
- MOB2 overexpression suppressed tumor growth in mouse xenograft models, with reductions in tumor volume up to 60% compared to vector controls [96].
- Analysis of TCGA data revealed MOB2 mRNA is significantly downregulated in GBM samples compared to low-grade gliomas (p = 3.94e-05) [96].
- Kaplan-Meier survival analysis demonstrated that low MOB2 expression correlates with poor prognosis in glioma patients (p = 0.00999) [96].
Table 2: Quantitative Effects of MOB2 Manipulation in Cancer Models
| Experimental Manipulation | Cancer Model | Observed Effects | Magnitude of Change |
|---|---|---|---|
| MOB2 Knockdown | Glioblastoma (LN-229, T98G cells) | Enhanced colony formation | 1.8-2.3 fold increase |
| MOB2 Knockdown | Glioblastoma (LN-229, T98G cells) | Increased migration | 2.1-2.7 fold increase |
| MOB2 Knockdown | Glioblastoma (LN-229, T98G cells) | Promoted invasion | 2.4-3.1 fold increase |
| MOB2 Overexpression | Glioblastoma (SF-539, SF-767 cells) | Suppressed colony formation | 2.1-2.8 fold decrease |
| MOB2 Overexpression | Chick CAM model | Reduced invasion | Significant decrease |
| MOB2 Overexpression | Mouse xenograft | Inhibited tumor growth | Up to 60% reduction |
MOB2 in DNA Damage Response and Cell Cycle Regulation
MOB2 plays a critical role in maintaining genome stability through its functions in DNA damage response:
- MOB2 depletion triggers a p53/p21-dependent G1/S cell cycle arrest in untransformed human cells [4].
- MOB2 knockdown causes accumulation of DNA damage and consequent activation of DDR kinases ATM and CHK2, even without exogenous DNA damage [4].
- MOB2 is required for cell survival and proper G1/S cell cycle arrest upon exposure to DNA damaging agents such as ionizing radiation or doxorubicin [4].
- MOB2 supports recruitment of MRN complex and activated ATM to DNA damaged chromatin, facilitating efficient DNA repair [4].
Experimental Protocols for Investigating MOB2 Specificity
Protein-Protein Interaction Mapping
Co-immunoprecipitation (Co-IP) and Western Blotting Purpose: To validate specific interactions between MOB2 and NDR1/2 kinases, and exclude interactions with LATS1/2. Methodology:
- Transfect cells with plasmids expressing tagged MOB2 (e.g., V5-MOB2) alongside FLAG-NDR1, FLAG-NDR2, FLAG-LATS1, or FLAG-LATS2.
- After 24-48 hours, lyse cells in NP-40 lysis buffer supplemented with protease and phosphatase inhibitors.
- Incubate cell lysates with anti-FLAG M2 affinity gel for 4 hours at 4°C.
- Wash beads extensively with lysis buffer, elute proteins with 2× Laemmli buffer, and separate by SDS-PAGE.
- Transfer to PVDF membrane and probe with anti-V5 and anti-FLAG antibodies. Expected Results: MOB2 should co-precipitate with NDR1 and NDR2, but not with LATS1 or LATS2 [4].
Yeast Two-Hybrid Screening Purpose: To identify novel binding partners of MOB2 beyond NDR kinases. Methodology:
- Clone MOB2 into pGBKT7 (DNA-BD vector) as bait.
- Screen against a human cDNA library cloned into pGADT7 (AD vector).
- Plate transformations on SD/-Leu/-Trp/-His/-Ade medium to select for protein interactions.
- Sequence positive clones and validate interactions by co-IP in mammalian cells. Application: This approach identified RAD50 as a novel MOB2 binding partner, linking MOB2 to DNA damage response [4].
Functional Characterization of MOB2-NDR Signaling
Kinase Activity Assays Purpose: To determine how MOB2 binding affects NDR1/2 kinase activity. Methodology:
- Purify recombinant NDR2, MOB1, and MOB2 proteins from E. coli or insect cells.
- Perform in vitro kinase reactions with NDR2 pre-incubated with either MOB1 or MOB2.
- Use myelin basic protein (MBP) or a specific NDR substrate peptide as phosphorylation substrate.
- Measure kinase activity by incorporation of 32P from [γ-32P]ATP or using phospho-specific antibodies. Expected Results: MOB1 should enhance NDR2 kinase activity, while MOB2 should suppress it [4].
Cell Cycle Analysis After MOB2 Depletion Purpose: To assess the role of MOB2 in cell cycle progression and DNA damage checkpoints. Methodology:
- Knock down MOB2 in untransformed human cells using specific shRNAs or siRNAs.
- Analyze cell cycle distribution by flow cytometry after propidium iodide staining.
- Monitor activation of cell cycle checkpoints by Western blotting for p53, p21, phospho-CHK1, and phospho-CHK2.
- Assess DNA damage accumulation by immunofluorescence for γH2AX foci. Expected Results: MOB2 depletion should cause G1/S arrest with increased p53/p21 expression and γH2AX foci [4].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Investigating MOB2 Specificity and Function
| Reagent Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Expression Plasmids | pCDH-MOB2-V5, FLAG-NDR1/2, FLAG-LATS1/2 | Protein interaction studies, functional rescue experiments | Include MOB2-H157A mutant defective in NDR binding as negative control [96] |
| Knockdown Tools | shRNA targeting MOB2 (e.g., TRCN0000195484), non-targeting shRNA control | Loss-of-function studies, phenotype characterization | Validate knockdown efficiency by Western blot; use multiple shRNAs to rule off-target effects [96] |
| Antibodies | Anti-MOB2 (e.g., Abcam ab230356), Anti-NDR1/2, Anti-LATS1/2, Anti-YAP/TAZ, Anti-pYAP-S127 | Protein detection, localization, activity assessment | Verify specificity using knockout/knockdown controls; phospho-antibodies require proper validation [96] |
| Cell Lines | LN-229, T98G (high endogenous MOB2), SF-539, SF-767 (low endogenous MOB2) | Disease modeling, functional assays | Use multiple cell lines to ensure generalizability; authenticate regularly [96] |
| Kinase Assay Components | Recombinant NDR2, MOB1, MOB2 proteins, [γ-32P]ATP, kinase buffer | In vitro kinase activity measurements | Include both active and kinase-dead NDR2 controls; optimize protein concentrations [4] |
Visualization of Signaling Pathways and Experimental Workflows
MOB2-NDR and MOB1-LATS Signaling Divergence
Experimental Workflow for MOB2 Functional Characterization
The specificity of MOB2 for NDR kinases establishes a critical regulatory branch point in the broader Hippo signaling network, creating a parallel pathway that operates alongside the canonical MOB1-LATS-YAP/TAZ axis. While MOB1 primarily regulates tissue growth and organ size through LATS-mediated control of YAP/TAZ, MOB2-NDR signaling modulates distinct cellular processes including cell cycle progression, DNA damage response, and cytoskeletal organization [96] [4]. This functional specialization highlights the evolutionary diversification of MOB family proteins as key determinants of signaling specificity within the NDR/LATS kinase family.
Future research should focus on elucidating the structural basis of MOB2-NDR specificity, particularly the molecular determinants that prevent MOB2 from interacting with LATS kinases despite their structural similarities to NDR kinases. Additionally, the context-dependent regulation of MOB2 expression and function in different tissue types and disease states remains poorly understood. The development of selective small molecule inhibitors or activators targeting the MOB2-NDR interface would provide valuable tools for dissecting this pathway's functions and exploring its therapeutic potential. As research progresses, the MOB2-NDR signaling axis may emerge as a promising target for therapeutic intervention in cancers where this pathway is dysregulated, particularly in glioblastoma and other malignancies characterized by MOB2 downregulation [96].
The MOB-NDR kinase signaling axis represents a critical regulatory node in cellular homeostasis, with profound implications for human disease pathogenesis. These evolutionarily conserved signaling pathways, central to the Hippo network, govern essential processes including cell proliferation, apoptosis, centrosome duplication, and morphogenesis [16] [2]. The specific binding partnerships between MOB coactivators and NDR/LATS kinases form distinct signaling modules that regulate diverse cellular outcomes. Understanding the structural and functional dichotomy between MOB1 and MOB2 binding specificity for NDR kinases provides a foundation for therapeutic intervention in cancer, neurological disorders, and developmental diseases. This technical guide examines the molecular basis of MOB-NDR interactions and explores emerging strategies for targeting these interfaces, with particular emphasis on exploiting the competitive binding relationship between MOB1 and MOB2 for NDR kinases as a novel therapeutic approach.
Table 1: Human MOB Protein Family Characteristics
| MOB Protein | Binding Partners | Functional Role | Regulatory Effect |
|---|---|---|---|
| MOB1A/B | NDR1/2, LATS1/2 | Tumor suppressor | Kinase activation [16] |
| MOB2 | NDR1/2 | Negative regulator | Kinase inhibition [16] |
| MOB3A/B/C | None identified | Unknown | No kinase binding or activation [16] |
MOB-NDR Biology and Binding Specificity
The MOB Protein Family and NDR Kinase Network
The human MOB protein family comprises six distinct members (MOB1A, MOB1B, MOB2, MOB3A, MOB3B, and MOB3C) that function as integral components of signaling pathways controlling critical cellular processes [16]. MOB proteins associate with and regulate the NDR/LATS kinases, which belong to the AGC family of serine-threonine kinases. The four human NDR/LATS kinases (NDR1, NDR2, LATS1, LATS2) share structural homology but exhibit distinct subcellular localization and functions [3]. NDR1 localizes to the nucleus, while NDR2 displays a punctate cytoplasmic distribution, suggesting divergent biological roles [3]. The MOB-NDR interaction is essential for kinase activity and function, with different MOB proteins exhibiting specific binding preferences. MOB1 proteins bind to and activate both NDR1/2 and LATS1/2 kinases, while MOB2 specifically interacts with NDR1/2 but not LATS kinases [16] [3].
Structural Basis of MOB-NDR Interactions
The molecular mechanisms governing MOB-NDR binding specificity are rooted in conserved structural features. All NDR/LATS kinases contain a characteristic N-terminal regulatory (NTR) region that binds specific MOB cofactors [1]. Structural analyses reveal that LATS kinases associate with MOB1 proteins, while NDR kinases preferentially bind MOB2 proteins [1]. The NDR/LATS NTR-Mob interface constitutes a structural platform that mediates kinase-cofactor binding and organizes the NTR to interact with the AGC kinase C-terminal hydrophobic motif (HM), facilitating allosteric regulation [1].
High-resolution crystal structures of Saccharomyces cerevisiae Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes demonstrate that the NTR forms a V-shaped helical hairpin similar to those observed in human Lats1–Mob1 and Ndr–Mob1 complexes [1]. This conserved architecture positions the phosphorylated threonine in the kinase's HM proximal to a conserved arginine residue in the NTR upon MOB binding, driving optimal orientation of the kinase's αC helix, a component critical for kinase activation [1]. Specificity determinants are restricted to discrete sites rather than broadly distributed across the interface, with a short motif in the MOB structure differing between MOB1 and MOB2 strongly contributing to molecular recognition [1].
Diagram 1: MOB-NDR/LATS Signaling Network. MOB1 activates both NDR and LATS kinases, while MOB2 competitively inhibits NDR kinase activation. These pathways regulate diverse cellular processes including proliferation, apoptosis, centrosome duplication, and neuronal remodeling.
Therapeutic Implications of MOB-NDR Interfaces
MOB2 as a Negative Regulator of NDR Kinases
A pivotal finding with significant therapeutic implications is the antagonistic relationship between MOB1 and MOB2 in regulating NDR kinase activity. Research demonstrates that MOB2 competes with MOB1A for NDR binding but functions as a negative regulator of NDR kinases in biochemical and biological settings [16]. Unlike MOB1A/B, which bind to and activate phosphorylated NDR, MOB2 associates specifically with unphosphorylated NDR and does not stimulate kinase activity [16]. This competitive binding has functional consequences, as RNA interference-mediated depletion of MOB2 results in increased NDR kinase activity, while MOB2 overexpression interferes with NDR roles in death receptor signaling and centrosome overduplication [16].
The competitive inhibition mechanism presents a novel approach for therapeutic intervention in diseases driven by aberrant NDR signaling. By modulating the MOB1/MOB2 balance, it may be possible to fine-tune NDR kinase activity for therapeutic benefit. In cancer, where NDR kinases can function as tumor suppressors, strategies to enhance MOB1 binding or disrupt MOB2-NDR interactions could restore growth control and promote apoptosis.
MOB-NDR Signaling in Neuronal Remodeling
Emerging evidence implicates MOB-NDR complexes in neuronal development and plasticity, suggesting therapeutic potential for neurological disorders. In C. elegans, the NDR kinase SAX-1 functions with its conserved interactors SAX-2/Furry and MOB-2 to control dendrite branch-specific elimination during stress-induced neuronal remodeling [17]. SAX-1/NDR promotes elimination of secondary and tertiary dendrites but not quaternary dendrites, demonstrating unexpected specificity in pruning processes [17]. This pathway functions with RABI-1/Rabin8 and the small GTPase RAB-11.2 to mediate endocytosis during remodeling, linking NDR signaling to membrane trafficking [17].
The conservation of this regulatory system in mammals suggests that targeting MOB-NDR interfaces could modulate neuronal connectivity in contexts of neurodevelopmental disorders, neurodegenerative diseases, or neural injury. The branch-specific effects of NDR signaling indicate particularly refined potential for interventions seeking to remodel specific neuronal connections without globally affecting dendritic architecture.
Table 2: Disease-Relevant Functions of MOB-NDR Complexes
| Disease Context | MOB-NDR Component | Functional Role | Therapeutic Implication |
|---|---|---|---|
| Cancer | MOB1A/B | Tumor suppressor via NDR/LATS activation | Enhance MOB1 function or expression [16] |
| Cancer | MOB2 | Negative regulator of NDR kinases | Inhibit MOB2-NDR interaction [16] |
| Neurological Disorders | SAX-1/NDR with MOB-2 | Dendrite pruning and neuronal remodeling | Modulate synaptic connectivity [17] |
| Fungal Pathogenesis | MOB1-DBF2, MOB2-COT1 | Septation and polar tip growth | Antifungal drug development [49] |
Experimental Approaches for Targeting MOB-NDR Interfaces
Structural Biology and Biophysical Methods
Elucidating the atomic-level details of MOB-NDR interactions provides the foundation for rational drug design. High-resolution X-ray crystallography has been instrumental in characterizing the Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes [1]. The experimental workflow involves:
Protein Expression and Purification: Recombinant expression of NDR N-terminal regulatory regions and MOB proteins in E. coli using expression vectors such as pGEX-4T1 or pMal-2c [16] [1]. MOB proteins often require stabilization strategies, such as engineering zinc-binding motifs (e.g., Mob2 V148C Y153C) to enable suitable expression for structural studies [1].
Crystallization and Structure Determination: Crystallization of MOB-NDR complexes using vapor diffusion methods. Data collection at synchrotron sources (e.g., wavelength ~0.978-1.000Å) followed by structure solution using molecular replacement [1]. Table 1 in [1] provides detailed crystallographic statistics including resolution, space group, cell dimensions, and refinement parameters.
Binding Affinity and Specificity Analysis: Surface plasmon resonance (SPR) or isothermal titration calorimetry (ITC) to quantify binding affinities and thermodynamic parameters of MOB-NDR interactions. Mutational analysis of interface residues to determine specificity determinants [1].
Cell-Based Functional Assays
Cell-based approaches validate the functional consequences of modulating MOB-NDR interfaces:
Interaction Mapping: Co-immunoprecipitation experiments in mammalian cells (e.g., HEK 293, HeLa) transfected with epitope-tagged (HA, myc) MOB and NDR constructs [16] [3]. Assessment of binding dependencies through deletion mutants and site-directed mutagenesis.
Kinase Activity Assays: Measurement of NDR kinase activity in response to MOB coexpression using immunocomplex kinase assays with appropriate substrates [16] [3]. Monitoring NDR autophosphorylation and phosphorylation of the hydrophobic motif by upstream kinases (e.g., MST1) [16].
Functional Consequences: RNA interference-mediated depletion (using pTER-shRNA vectors) or overexpression of MOB proteins to assess effects on NDR-dependent processes including apoptosis, centrosome duplication, and morphological changes [16]. For neuronal remodeling studies, in vivo analysis in model organisms like C. elegans using specific neuronal markers [17].
Diagram 2: Experimental Approach for MOB-NDR Interface Studies. Integrated methodologies spanning structural biology, biophysical analysis, cellular assays, and functional validation provide comprehensive characterization of MOB-NDR interactions and their therapeutic potential.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for MOB-NDR Investigations
| Reagent/Tool | Specifications | Experimental Application | Key References |
|---|---|---|---|
| Expression Vectors | pcDNA3, pGEX-4T1, pMal-2c with BamHI/XhoI sites | Mammalian and bacterial expression of epitope-tagged MOB/NDR proteins | [16] |
| MOB/NDR cDNA Constructs | Human: MOB1A, MOB1B, MOB2, MOB3A-C, NDR1, NDR2, LATS1, LATS2 | Interaction mapping and functional studies | [16] [3] |
| RNAi Vectors | pTER-shMOB2 with BglII/HindIII sites | Knockdown of specific MOB proteins | [16] |
| Cell Lines | COS-7, HEK 293, U2-OS, HeLa, Jurkat T-cells | Protein interaction, localization, and kinase activity assays | [16] [3] |
| Crystallography Reagents | Zinc-binding Mob2 (V148C Y153C), Cbk1NTR (251-351) | Structural studies of MOB-NDR complexes | [1] |
| C. elegans Strains | IL2 neuron markers, sax-1, mob-2 mutants | Neuronal remodeling studies | [17] |
The MOB-NDR interface represents a promising but underexplored therapeutic target with broad relevance to cancer, neurological disorders, and other diseases. The precise binding specificity between MOB coactivators and NDR kinases, particularly the competitive relationship between MOB1 and MOB2, offers multiple avenues for therapeutic intervention. Future directions should include high-throughput screening for small molecules that modulate MOB-NDR interactions, development of peptide-based inhibitors targeting the specific binding interfaces, and gene therapy approaches to manipulate the MOB1/MOB2 balance in disease contexts. The conserved nature of these signaling modules across evolution underscores their fundamental importance in cellular regulation and enhances the translational potential of targeting these interfaces for therapeutic benefit.
Conclusion
The specific binding relationships between MOB proteins and NDR kinases represent a sophisticated regulatory mechanism within the Hippo signaling network. Structural studies reveal that discrete molecular interfaces, rather than broadly distributed regions, determine the precise specificity of MOB1 for LATS and MOB2 for NDR kinases. The functional relationship between MOB2 and NDR is complex and context-dependent, with MOB2 acting not merely as an inhibitor but as a nuanced regulator that competes with MOB1 and influences downstream effectors including YAP/TAZ. These interactions have profound implications for cell cycle control, DNA damage response, and cell motility, particularly in cancer pathogenesis. Future research should focus on developing small molecule modulators of these specific interactions, exploring tissue-specific variations in MOB-NDR signaling, and investigating the therapeutic potential of targeting these interfaces in cancer and other diseases. The integrated understanding of MOB-NDR specificity provides a foundation for novel therapeutic strategies that could precisely modulate Hippo pathway signaling.
References
- 1. Ndr/Lats Kinases Bind Specific Mob-Family Coactivators ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8341033/]
- 2. The Structure of an NDR/LATS Kinase–Mob Complex Reveals ... [https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1002146]
- 3. Human Mob Proteins Regulate the NDR1 and NDR2 ... [https://www.sciencedirect.com/science/article/pii/S0021925820665796]
- 4. The Possible Crosstalk of MOB2 With NDR1/2 Kinases in Cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5321467/]
- 5. hippo [https://www.sdbonline.org/sites/fly/sturtevant/hippo1.htm]
- 6. The NDR/LATS protein kinases in neurobiology [https://www.sciencedirect.com/science/article/pii/S0171933523000481]
- 7. Mapping the MOB proteins' proximity network reveals a ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10480535/]
- 8. NDR2 kinase: A review of its physiological role and ... [https://www.sciencedirect.com/science/article/pii/S0141813025042084]
- 9. The NDR family of kinases: essential regulators of aging [https://www.frontiersin.org/journals/molecular-neuroscience/articles/10.3389/fnmol.2024.1371086/full]
- 10. Mechanism of Activation of NDR (Nuclear Dbf2-related) ... [https://www.sciencedirect.com/science/article/pii/S0021925820730991]
- 11. CDK-dependent phosphorylation of Mob2 is essential for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3135472/]
- 12. The NDR/LATS Family Kinase Cbk1 Directly Controls ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2517623/]
- 13. Human NDR Kinases Are Rapidly Activated by MOB Proteins ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1234321/]
- 14. The Structure of an NDR/LATS Kinase–Mob Complex Reveals ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4428629/]
- 15. Mob Family Proteins: Regulatory Partners in Hippo and ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2020.00161/full]
- 16. Differential NDR/LATS Interactions with the Human MOB ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2937529/]
- 17. NDR kinase SAX-1 controls dendrite branch-specific ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12259037/]
- 18. Stable MOB1 interaction with Hippo/MST is not essential for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5612953/]
- 19. MOB1 Mediated Phospho-recognition in the Core ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5461540/]
- 20. Regulation of Protein Interactions by Mps One Binder ... [https://www.sciencedirect.com/science/article/pii/S1535947620323732]
- 21. The LATS1 and LATS2 tumor suppressors [https://www.nature.com/articles/cdd201799]
- 22. Stable MOB1 interaction with Hippo/MST is not essential ... [https://www.nature.com/articles/s41467-017-00795-y]
- 23. Monopolar spindle-one-binder protein 2 regulates the ... [https://www.spandidos-publications.com/10.3892/ol.2018.7952]
- 24. Structural Basis for Auto-Inhibition of the NDR1 Kinase ... [https://www.sciencedirect.com/science/article/pii/S0969212618301783]
- 25. 5XQZ: Structure of the MOB1-NDR2 complex [https://www.rcsb.org/structure/5xqz]
- 26. Regulation of NDR Protein Kinase by Hydrophobic Motif ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1316964/]
- 27. Structural insights of AKT and its activation mechanism for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12638386/]
- 28. Regulation of NDR2 Protein Kinase by Multi-site ... [https://www.sciencedirect.com/science/article/pii/S002192582066759X]
- 29. Regulation of NDR Protein Kinase by Hydrophobic Motif ... [https://pubmed.ncbi.nlm.nih.gov/16314523/]
- 30. mediated regulation of NDR protein kinase through ... [https://www.sciencedirect.com/science/article/pii/S0021925819325918]
- 31. Computational drug development for membrane protein ... [https://pubmed.ncbi.nlm.nih.gov/38361054/]
- 32. The role of structural biology in drug discovery [https://www.jeolusa.com/NEWS-EVENTS/Blog/the-role-of-structural-biology-in-drug-discovery]
- 33. Structural basis for autoinhibition and its relief of MOB1 in ... [https://www.nature.com/articles/srep28488]
- 34. Cryo electron microscopy to determine the structure of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5405050/]
- 35. Structural determination of small proteins by cryo-EM using ... [https://www.nature.com/articles/s41598-025-20608-3]
- 36. Article Crystal Structure of a Human Mob1 Protein [https://www.sciencedirect.com/science/article/pii/S0969212603001825]
- 37. Sequence and Structure-Based Analysis of Specificity ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7809594/]
- 38. Regulation of Protein Interactions by Mps One Binder (MOB1 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5461541/]
- 39. Co-immunoprecipitation (Co-IP): The Complete Guide [https://www.antibodies.com/applications/co-immunoprecipitation]
- 40. Co-immunoprecipitation: Principles and applications [https://www.abcam.com/en-us/knowledge-center/proteins-and-protein-analysis/co-immunoprecipitation]
- 41. The value of coimmunoprecipitation (Co-IP) assays in drug ... [https://pubmed.ncbi.nlm.nih.gov/40289752/]
- 42. Co-Immunoprecipitation (Co-IP) and Pull-Down [https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-assays-analysis/protein-interaction-analysis/co-immunoprecipitation-co-ip-pull-down.html]
- 43. Co-immunoprecipitation and semi-quantitative ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8264609/]
- 44. Mechanism of activation of NDR (nuclear Dbf2-related) ... [https://pubmed.ncbi.nlm.nih.gov/15197186/]
- 45. Choosing the Right Assay for Your Kinase Drug Discovery [https://www.reactionbiology.com/choosing-the-right-assay-for-your-kinase-drug-discovery/]
- 46. Comprehensive assay of kinase catalytic activity reveals ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3230241/]
- 47. RNAi vs. CRISPR: Guide to Selecting the Best Gene ... [https://www.synthego.com/blog/rnai-vs-crispr-guide/]
- 48. The Researcher's Guide to Designing CRISPR and RNAi ... [https://www.merckmillipore.com/CI/fr/technical-documents/technical-article/genomics/functional-genomics-screening/crispr-rnai-screening-guide]
- 49. Two NDR kinase–MOB complexes function as distinct ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4617822/]
- 50. Genome-wide RNA interference screening reveals a COPI ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7443978/]
- 51. RNAi-mediated control of CRISPR functions - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7295050/]
- 52. Ndr kinases regulate retinal interneuron proliferation and ... [https://www.nature.com/articles/s41598-018-30492-9]
- 53. Furry protein suppresses nuclear localization of yes ... [https://www.sciencedirect.com/science/article/pii/S002192581749507X]
- 54. Control of stress-activated Cdc42 dynamics by the MAP ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12397940/]
- 55. Unveiling the hidden interactome of CRBN molecular glues [https://www.nature.com/articles/s41467-025-62099-w]
- 56. The SpyBLI cell-free pipeline for the rapid quantification of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12247212/]
- 57. Protein kinases in neurodegenerative diseases [https://www.nature.com/articles/s41392-025-02179-x]
- 58. Phosphorylation State-Specific Antibodies - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC1892416/]
- 59. Characterization and Evolution of the Cell Cycle-Associated ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2684140/]
- 60. Phospho-Specific Antibodies for Flow Cytometry [https://www.thermofisher.com/us/en/home/life-science/cell-analysis/flow-cytometry/antibodies-for-flow-cytometry/phospho-specific-antibodies-flow-cytometry.html]
- 61. Overcoming non-specific binding to measure the active ... [https://www.sciencedirect.com/science/article/abs/pii/S0956566318304433]
- 62. Overcoming nonspecific binding challenges in PK assays [https://labtesting.wuxiapptec.com/2024/12/26/overcoming-nonspecific-binding-challenges-in-pk-assays/]
- 63. A Method for Removing Effects of Nonspecific Binding on ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2931719/]
- 64. NDR2 Kinase Regulates Microglial Metabolic Adaptation and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12609617/]
- 65. Chemical genetic identification of NDR1/2 kinase substrates ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3333840/]
- 66. The Roles of NDR Protein Kinases in Hippo Signalling [https://www.mdpi.com/2073-4425/7/5/21]
- 67. The NDR family of kinases: essential regulators of aging - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11129689/]
- 68. Quantitative maps of protein phosphorylation sites across ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3621391/]
- 69. The Roles of NDR Protein Kinases in Hippo Signalling - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4880841/]
- 70. Monopolar spindle-one-binder protein 2 regulates the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5840656/]
- 71. Overview of methods for comparing the efficacies of drugs ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3895352/]
- 72. Table 3, Summary of headings for reporting the quantitative ... [https://www.ncbi.nlm.nih.gov/books/NBK132268/table/cerguideguidance.t3/]
- 73. Crosstalk between NDR kinase pathways coordinates cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3224761/]
- 74. Analysis of intracellular and intercellular crosstalk from omics ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0334981]
- 75. Mob Family Proteins: Regulatory Partners in Hippo and Hippo ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7096357/]
- 76. Mob2 Insufficiency Disrupts Neuronal Migration in the ... [https://www.frontiersin.org/journals/cellular-neuroscience/articles/10.3389/fncel.2018.00057/full]
- 77. Identification of Mob2, a Novel Regulator of Larval ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3813873/]
- 78. "MOB-NDR Kinase Signaling Components are Required for ... [https://oasis.library.unlv.edu/thesesdissertations/3486/]
- 79. Growth-inhibitory effects of MOB2 on human hepatic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3544031/]
- 80. The Problem of Partial Competition in the Quantitative ... [https://www.sciencedirect.com/science/article/abs/pii/S0003269783713928]
- 81. Generating quantitative binding landscapes through ... [https://www.nature.com/articles/s41467-019-13895-8]
- 82. Regulation of NDR1 activity by PLK1 ensures proper ... [https://www.nature.com/articles/srep10449]
- 83. Experimental Models for Preclinical Research in ... - NCBI [https://www.ncbi.nlm.nih.gov/books/NBK553756/]
- 84. Preclinical Models of Hepatocellular Carcinoma [https://www.mdpi.com/2227-9059/12/7/1624]
- 85. Hepatocellular carcinoma: signaling pathways and ... [https://www.nature.com/articles/s41392-024-02075-w]
- 86. Exploring the role of the immune microenvironment in ... [https://elifesciences.org/articles/95009]
- 87. Regulation of DNA damage responses and cell cycle ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4276419/]
- 88. Regulation of DNA damage responses and cell cycle ... [https://www.sciencedirect.com/science/article/abs/pii/S0898656814003702]
- 89. Human NDR Kinases Control G1/S Cell Cycle Transition ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3135299/]
- 90. Addressing cancer invasion and cell motility with ... [https://www.nature.com/articles/s41598-022-05307-7]
- 91. Functional assessment of migration and adhesion to quantify ... [https://pubs.rsc.org/en/content/articlehtml/2025/sm/d4sm01351d]
- 92. Quantitative impedance-based characterization of breast ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12512568/]
- 93. Quantitative cell imaging approaches to metastatic state ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2022.1048630/full]
- 94. The components and regulation of the Hippo pathway and its ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12366177/]
- 95. The Hippo signalling pathway and its implications in ... [https://www.nature.com/articles/s41392-022-01191-9]
- 96. MOB2 suppresses GBM cell migration and invasion via ... [https://www.nature.com/articles/s41419-020-2381-8]
- 97. Hippo/YAP signaling's multifaceted crosstalk in cancer [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1595362/full]
- 98. The multifaceted role of YAP in the tumor ... [https://www.nature.com/articles/s12276-025-01551-9]
- 99. YAP/TAZ: An epitome of tumorigenesis [https://www.sciencedirect.com/science/article/abs/pii/S0304383525003738]