Hydrogen Bonding in Primer Dimers: A Scientific Guide to Mechanisms, Detection, and Prevention
This article provides a comprehensive analysis of the critical role hydrogen bonding plays in the formation and prevention of primer dimers, a common obstacle in molecular biology and diagnostic assay...
Hydrogen Bonding in Primer Dimers: A Scientific Guide to Mechanisms, Detection, and Prevention
Abstract
This article provides a comprehensive analysis of the critical role hydrogen bonding plays in the formation and prevention of primer dimers, a common obstacle in molecular biology and diagnostic assay development. Tailored for researchers, scientists, and drug development professionals, the content explores the fundamental biophysical principles governing these non-specific interactions. It further details advanced methodological strategies for detection and mitigation, presents comparative data on troubleshooting and optimization techniques, and validates innovative approaches like Self-Avoiding Molecular Recognition Systems (SAMRS). By synthesizing foundational knowledge with practical applications, this guide aims to empower professionals in designing more robust and reliable molecular assays.
The Hydrogen Bond Blueprint: Deconstructing the Biophysics of Primer Dimers
In nucleic acid amplification technologies, primer dimers represent a significant challenge to assay specificity and efficiency. These unintended artifacts are short, aberrant DNA fragments generated when primers anneal to each other rather than to the target template DNA, becoming substrates for polymerase-mediated extension [1]. The formation of these structures is governed fundamentally by Watson-Crick hydrogen bonding between complementary bases, the same force that facilitates specific primer-template interactions [2]. While high-fidelity replicative DNA polymerases rely primarily on geometric constraints for fidelity, studies demonstrate that Y-family polymerases involved in lesion bypass depend critically on Watson-Crick hydrogen bonding to localize nascent base pairs in their active sites [2]. This dependency underscores the dual role of hydrogen bonding in molecular biology: it is essential for desired specific amplification yet equally capable of facilitating undesirable primer-primer interactions when complementarity exists. This technical guide examines the classification, formation mechanisms, detection, and mitigation of primer dimers within the broader context of hydrogen bonding energetics in nucleic acid biochemistry.
Classification and Formation Mechanisms
Structural Classification of Primer Dimers
Primer dimers are systematically categorized based on the interacting primers involved:
Self-Dimers (Homodimers): Formed when two identical primers anneal to each other through intermolecular bonding [3]. This occurs when a single primer sequence contains regions of self-complementarity.
Cross-Dimers (Heterodimers): Formed when forward and reverse primers with complementary sequences anneal to each other instead of the target template [1] [3].
The thermodynamic driving force for dimer formation is the Gibbs free energy (ΔG) released when complementary sequences hybridize. More negative ΔG values indicate stronger, more stable dimer formations that are increasingly problematic in amplification reactions [4].
Molecular Mechanisms of Dimer Formation
The process of primer dimer formation follows a predictable pathway, illustrated below, which initiates with hydrogen bonding between complementary primer regions and culminates in polymerase-mediated extension:
The diagram illustrates the two-phase process of primer dimer formation. The initial hydrogen bonding phase relies on Watson-Crick base pairing between complementary regions of primers, typically involving 3 or more complementary bases [4]. This creates short double-stranded regions with free 3' hydroxyl ends. During the subsequent polymerase extension phase, DNA polymerase recognizes these free 3' ends as legitimate initiation points and extends the primers, effectively "cementing" the dimer into a stable, amplifiable product [1] [3]. This extension process is particularly efficient when complementarity occurs at the 3' ends of primers, where polymerase binding initiates [3].
Quantitative Impact on Assay Performance
Effects on PCR and qPCR Efficiency
Primer dimers impact molecular assays through multiple mechanisms, with particularly severe consequences in quantitative applications:
Reduced Amplification Efficiency: Primer dimers sequester primers into non-productive complexes, effectively reducing the concentration of primers available for target amplification [5]. This leads to diminished target amplicon yield and reduced assay sensitivity.
qPCR Artifacts: In quantitative PCR using intercalating dyes, primer dimers generate false fluorescent signals as the dyes bind to double-stranded dimer products [6]. This is particularly problematic during later amplification cycles, potentially leading to inaccurate quantification [7].
Inhibition of Target Amplification: The presence of amplifiable primer dimers creates competition for essential reaction components, including primers, nucleotides, and DNA polymerase [3]. This resource partitioning can completely suppress target amplification in severe cases.
The table below summarizes the quantitative impacts of primer dimers on key assay parameters:
Table 1: Quantitative Impact of Primer Dimers on Assay Performance
| Assay Parameter | Impact of Primer Dimers | Experimental Manifestation | Reference |
|---|---|---|---|
| Amplification Efficiency | Decreased by 10-30% | Higher Cq values, reduced slope in standard curve | [7] |
| Detection Sensitivity | 1-3 log10 reduction in sensitivity | Increased limit of detection | [5] |
| Reaction Resources | Up to 50% primer sequestration | Reduced yield of desired product | [1] |
| qPCR Accuracy | False positive signals, efficiency >100% | Incorrect quantification, elevated baseline | [7] |
Special Considerations for Isothermal Amplification
Loop-mediated isothermal amplification (LAMP) presents unique vulnerabilities to primer dimer formation due to its structural complexity:
- Increased Primer Load: LAMP utilizes 4-6 primers targeting 6-8 regions, substantially increasing the probability of primer-primer interactions [5].
- Complex Primer Structures: Inner primers (FIP/BIP) in LAMP are typically 40-45 bases long, making them particularly prone to stable hairpin formation and self-dimerization [5].
- High Primer Concentrations: Standard LAMP protocols use inner primer concentrations of 1.6 µM, approximately 10-fold higher than conventional PCR, increasing interaction probabilities [5].
Research demonstrates that primer sets with strong 3' complementarity can generate a slowly rising baseline in real-time LAMP assays due to amplifiable primer dimers and hairpin structures, significantly compromising endpoint detection clarity [5].
Detection and Characterization Methods
Electrophoretic and Analytical Techniques
Multiple experimental approaches enable detection and characterization of primer dimers:
Gel Electrophoresis: Primer dimers typically appear as fuzzy, smeary bands below 100 bp, distinct from the well-defined bands of specific amplicons [1]. Running gels for extended time helps separate primer dimers from desired products.
No-Template Controls (NTC): Essential diagnostic reactions containing all PCR components except template DNA. Amplification in NTCs indicates primer-derived artifacts rather than target-specific products [1].
Melting Curve Analysis: Following qPCR with intercalating dyes, melting curves reveal primer dimers through distinct melting temperatures (Tm) that are typically lower than specific amplicons [6].
Thermodynamic Prediction Tools: Software such as OligoAnalyzer and Multiple Prime Analyzer calculates ΔG values for potential dimer formations, with structures exceeding -9 kcal/mol considered problematic [8].
The experimental workflow below outlines a comprehensive approach to primer dimer detection and validation:
This workflow emphasizes the importance of combining computational prediction with experimental validation. The computational phase identifies potential dimerization risks through thermodynamic modeling, while the experimental phase confirms actual dimer formation under reaction conditions. Research demonstrates that primer sets passing in silico screening may still produce problematic dimers in amplification reactions, necessitating empirical validation [5].
Experimental Mitigation Strategies
Primer Design Optimization
Strategic primer design represents the most effective approach to minimizing primer dimer formation:
3' End Complementarity Analysis: Scrutinize the last 3-5 bases at the 3' end for complementarity between primers, as this region most significantly influences dimer formation [4].
Gibbs Free Energy Thresholds: Avoid primer pairs with ΔG values below -9 kcal/mol for cross-dimers and below -2 kcal/mol for 3' end hairpins [8] [4].
GC Clamp Implementation: Include 1-2 G or C bases at the 3' end to enhance specific binding, but avoid more than 3 G/C in the final five bases to prevent non-specific priming [8].
Length Optimization: Design primers between 18-24 nucleotides to balance specificity and binding efficiency while minimizing secondary structure risk [8].
The table below outlines essential computational tools for primer design and dimer prediction:
Table 2: Computational Tools for Primer Dimer Prediction and Analysis
| Tool Name | Primary Function | Key Dimer-Related Parameters | Access Platform |
|---|---|---|---|
| Primer-BLAST | Integrated primer design with specificity checking | Off-target binding prediction, dimer flags | NCBI Web Portal |
| OligoAnalyzer | Thermodynamic analysis of oligonucleotides | ΔG calculation for dimers and hairpins | IDT Web Portal |
| Multiple Prime Analyzer | Multi-primer interaction analysis | Comprehensive dimer network prediction | Thermo Fisher Web Portal |
| mFold | Secondary structure prediction | Stability analysis of hairpin formations | Web-based application |
Reaction Condition Optimization
When primer redesign is not feasible, reaction condition adjustments can suppress dimer formation:
Hot-Start DNA Polymerase: Employ polymerases that remain inactive until elevated temperatures are reached, preventing primer dimer formation during reaction setup [1] [9].
Increased Annealing Temperature: Raise annealing temperature 2-5°C above the calculated Tm to reduce non-specific interactions while maintaining specific binding [1].
Primer Concentration Titration: Lower primer concentrations (50-200 nM for qPCR) to reduce interaction probabilities while maintaining amplification efficiency [1].
Magnesium Concentration Optimization: Titrate Mg²⁺ concentrations, as excess magnesium can stabilize non-specific primer interactions [3].
Recent research demonstrates that incorporating modified nucleotides such as digoxigenin-labeled dUTP can selectively prevent primer dimer detection in lateral flow assays, as dimers contain only one label and fail to generate signal despite their presence [10].
Research Reagent Solutions
The following reagents and tools represent essential components for effective primer dimer management in molecular assays:
Table 3: Essential Research Reagents for Primer Dimer Management
| Reagent/Tool | Specific Function | Dimer-Related Application | Example Products |
|---|---|---|---|
| Hot-Start DNA Polymerase | Thermal activation prevents pre-PCR activity | Suppresses dimer formation during reaction setup | Bst 2.0 WarmStart, Taq Hot Start |
| Thermodynamic Prediction Software | Calculates interaction energies | Identifies problematic primer pairs pre-synthesis | OligoAnalyzer, mFold |
| No-Template Control Reagents | Validates reaction specificity | Detects primer-derived amplification artifacts | Molecular biology grade water |
| Intercalating Dyes | Binds double-stranded DNA | Enables dimer detection in real-time and melt curves | SYTO 9, SYBR Green |
| Modified Nucleotides | Incorporates non-standard bases | Prevents dimer detection in endpoint assays | Digoxigenin-dUTP, Biotin-dATP |
Primer dimer formation represents a fundamental challenge in nucleic acid amplification technologies, rooted in the same Watson-Crick hydrogen bonding principles that enable specific target recognition. The competitive binding between desired primer-template interactions and undesirable primer-primer interactions directly impacts assay sensitivity, specificity, and quantification accuracy. Effective management requires integrated computational and experimental strategies, from careful primer design with thermodynamic considerations to optimized reaction conditions that favor specific amplification. As molecular diagnostics advance toward point-of-care applications and isothermal methods, understanding and controlling primer dimer formation becomes increasingly critical for assay reliability. The strategies outlined in this guide provide a framework for diagnosing and addressing primer dimer issues across various amplification platforms, emphasizing the continuous balance between harnessing and controlling hydrogen bonding energetics in molecular biology.
The specific pairing of nitrogenous bases—guanine with cytosine (G-C) and adenine with thymine (A-T) or uracil (A-U in RNA)—through hydrogen bonds constitutes a foundational principle of molecular biology [11] [12]. This complementary base pairing, first elucidated by Watson and Crick, is essential for the storage and replication of genetic information in DNA and RNA [13]. While the overall stability of the DNA double helix is significantly influenced by base-stacking interactions between adjacent nucleotide pairs, hydrogen bonding plays a critical role in defining the specificity and fidelity of base pairing [14] [11]. This specificity is not only crucial for in vivo processes like DNA replication and transcription but also forms the basis of numerous in vitro molecular techniques. Among these, the phenomenon of primer dimer formation in amplification reactions like PCR and LAMP represents a significant challenge where unintended hydrogen bonding between primers themselves, rather than with the target template, leads to non-specific amplification and potential false-positive results [5] [3]. This guide provides an in-depth technical examination of the hydrogen bonding patterns that govern natural base pairing, their quantitative energetic contributions, and their direct implications for the design and optimization of molecular assays, with a particular focus on troubleshooting primer dimer artifacts.
The Molecular Anatomy of Hydrogen Bonds in Base Pairs
Fundamental Hydrogen Bonding Patterns
Hydrogen bonds in base pairs form between hydrogen bond donors (atoms bearing a hydrogen, typically N-H or O-H groups) and hydrogen bond acceptors (electronegative atoms with lone electron pairs, such as nitrogen or oxygen) [15]. The specific patterns of donors and acceptors dictate which bases can form stable, complementary pairs.
- G-C Base Pair: The guanine-cytosine pair forms three hydrogen bonds. Guanine provides a hydrogen bond donor from its N1-H group and an acceptor from its O6 carbonyl group. It also provides an acceptor from the N7 position in its five-membered ring. Cytosine complements this with a donor from its N4-H₂ group and an acceptor from its N3 atom. The canonical pairing involves the N1-H⋯O2, N2-H⋯N3, and O6⋯H4-N4 interactions, creating a robust, thermodynamically stable pair [15] [11].
- A-T Base Pair: The adenine-thymine pair forms two hydrogen bonds. Adenine provides a donor from its N6-H₂ group and an acceptor from its N1 atom. Thymine complements this with an acceptor from its O4 carbonyl oxygen and a donor from its N3-H group. The standard pairing is N1⋯H-N3 and N6-H⋯O4 [15] [13].
The purine-pyrimidine pairing (A-T and G-C) is geometrically optimal. Pyrimidine-pyrimidine pairs are too short to bridge the inter-sugar distance, while purine-purine pairs are too long, leading to steric clash and inefficient overlap repulsion [11]. The distance from sugar linkage to sugar linkage is nearly identical for A-T and G-C pairs, allowing the DNA backbone to maintain a regular helical structure [13].
Table 1: Hydrogen Bond Donors and Acceptors in Natural Base Pairs
| Base | Hydrogen Bond Donors | Hydrogen Bond Acceptors | Complementary Base | Total H-Bonds |
|---|---|---|---|---|
| Guanine (G) | N1-H, N2-H₂ | O6, N7 | Cytosine (C) | 3 |
| Cytosine (C) | N4-H₂ | N3, O2 | Guanine (G) | 3 |
| Adenine (A) | N6-H₂ | N1, N3, N7 | Thymine (T) | 2 |
| Thymine (T) | N3-H | O2, O4 | Adenine (A) | 2 |
| Uracil (U) | N3-H | O2, O4 | Adenine (A) | 2 |
Relative Strength and Energetic Contributions
The stability of a base pair is directly related to its number of hydrogen bonds, with the G-C pair being stronger than the A-T pair. This difference is a primary reason why DNA stability and melting temperature (Tₘ) are dependent on GC content [11]. DNA with high GC-content is more stable and has a higher melting point than DNA with low GC-content.
However, the simplistic view that three hydrogen bonds automatically make G-C much stronger than A-T is nuanced by advanced computational studies. Quantum chemical analyses reveal that the intrinsic strength of individual hydrogen bonds varies. One study found that the most favorable hydrogen bond in both natural and unnatural base pairs is N-H⋯N, while O-H⋯N/O bonds are less favorable [16]. Furthermore, non-classical C-H⋯O/N bonds, particularly C-H⋯O bonds in Watson-Crick base pairs, play a significant and previously underappreciated role in stabilization [16]. When studied in a DNA environment using a QM/MM approach, the strength of the central N-H⋯N bond and the C-H⋯O bonds increases, while the strength of the N-H⋯O bond decreases, though the overall trends remain [16].
Critically, while hydrogen bonding is essential for specificity, π-π stacking interactions between adjacent base pairs in the double helix are primarily responsible for the overall stabilisation of the structure; the contribution of Watson-Crick base pairing to global structural stability is minimal in comparison to stacking [14] [11]. The interplay between these forces is complex, with studies showing that stacking can reinforce hydrogen bonding and vice versa, a phenomenon known as cooperativity [14].
Quantitative Analysis of Base Pair Stability
Thermodynamic Parameters
The stability of base pairs can be quantified using thermodynamic parameters derived from computational chemistry and experimental data. Energy decomposition analyses based on Kohn-Sham molecular orbital theory provide detailed insight into the various interactions that contribute to the stability of stacked base pairs in B-DNA [14]. These analyses break down the total interaction energy (ΔEint) into several components:
- ΔVelstat: The classical electrostatic interaction between the unperturbed charge distributions of the fragments.
- ΔEPauli: The Pauli repulsion, comprising destabilizing interactions between occupied orbitals, responsible for steric repulsion.
- ΔEoi: The orbital interaction, accounting for charge transfer and polarization.
- ΔEdisp: The dispersion interaction, accounting for correlation effects.
Table 2: Energy Decomposition Analysis (kcal mol⁻¹) for Selected Stacked Base Pairs (BP86-D/TZ2P level, Twist Angle = 36°)
| Stacked Base Pair System | ΔEint | ΔVelstat | ΔEPauli | ΔEoi | ΔEdisp | Hydrogen Bonding Contribution | Stacking Contribution |
|---|---|---|---|---|---|---|---|
| (G-C)/(G-C) | -64.5 | -90.1 | 117.0 | -49.7 | -41.7 | ~40% | ~60% |
| (A-T)/(A-T) | -41.2 | -63.8 | 94.2 | -32.9 | -38.7 | ~30% | ~70% |
| Mismatched Pair | ~ -20 to -35 | Varies | Varies | Varies | Varies | Lower | Lower |
This analysis reveals that stacking interactions (π-π) contribute more to the overall stability of the double helix than the hydrogen bonds within individual base pairs [14]. For instance, in the (G-C)/(G-C) stack, stacking can account for approximately 60% of the stabilisation. The twist angle between stacked base pairs is also a critical factor, with studies showing that twisting from 0° to the canonical 36° provides an additional stabilization of 6 to 12 kcal mol⁻¹ across different base pair stacks [14].
Impact of Solvation
The aqueous environment profoundly impacts hydrogen bonding. In the condensed phase (e.g., water), all hydrogen bonds of the base pairs become weaker and most bonds elongate [14]. This is because the lone pairs on atoms involved in hydrogen bonding are stabilized by the solvent, reducing their energy and availability for interaction. The solvent competes for hydrogen bonding sites, which can weaken the intramolecular base pairing. This desolvation penalty must be paid for hydrogen bonds to form in an aqueous environment, a key consideration for reactions like PCR that occur in solution [14].
Hydrogen Bonding in the Context of Primer Dimers
The Primer Dimer Problem
In molecular techniques such as the Polymerase Chain Reaction (PCR) and Loop-Mediated Isothermal Amplification (LAMP), synthetic oligonucleotide primers must bind specifically to a target DNA template. Primer dimers are artifacts formed when these primers bind to each other via complementary base pairing instead of to the target template [3]. This occurs due to unintended hydrogen bonding between primers.
- Homodimers vs. Heterodimers: A homodimer is formed when two identical primers bind, while a heterodimer is formed when forward and reverse primers with complementary sequences bind [3].
- Consequences: Once formed, primer dimers can be extended by DNA polymerase, leading to nonspecific amplification. This depletes reaction reagents (primers, nucleotides, enzyme) and generates non-target products, which can cause false-positive signals, reduce amplification efficiency, and lower assay sensitivity [5] [3].
The problem is exacerbated in techniques like LAMP, which uses 4-6 primers targeting 6-8 regions simultaneously. The high primer concentration and the long length of inner primers (FIP and BIP, typically 40-45 bases) increase the probability of primer-primer hybridization and the formation of stable secondary structures like hairpins [5] [3].
Hydrogen Bonding as the Root Cause
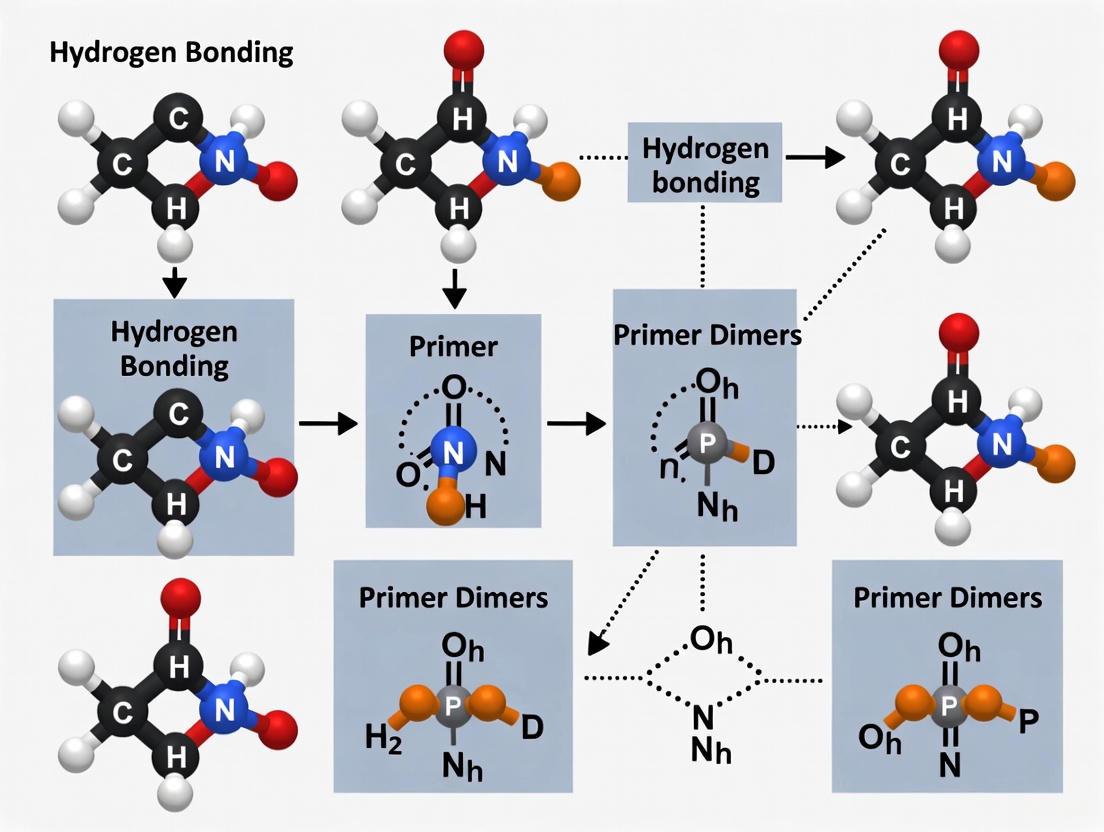
The formation of primer dimers is a direct consequence of predictable hydrogen bonding between short, complementary sequences within the primers themselves.
- 3'-End Complementarity: If the 3' ends of two primers have complementary sequences, even as short as 2-3 bases, they can anneal. DNA polymerase can then bind and extend the annealed primers in both directions, creating a short, double-stranded "primer dimer" product that amplifies efficiently [3] [17].
- Inter-Primer Homology: Complementary sequences anywhere within the primers can lead to hybridization. If the 3' ends are involved, extension is efficient; if binding occurs at the 5' end or middle, the polymerase may not extend efficiently, but the hybridized primers are still sequestered and unable to participate in target amplification [3] [17].
- GC Clamps and Repetitive Sequences: Primers with high GC content, especially at the 3' end (a design feature sometimes used to enhance binding, known as a GC clamp), have stronger hydrogen bonding. This can be beneficial for specific binding but also increases the risk of dimer formation if complementarity exists. Similarly, runs of a single base (e.g., AAAA or CCCC) or dinucleotide repeats (e.g., ATATAT) increase the chance of mispriming [17].
Experimental Protocols and Mitigation Strategies
In Silico Primer Analysis and Design
Preventing primer dimers begins with careful primer design and analysis using thermodynamic tools.
Protocol: Analyzing Primers for Secondary Structures
- Sequence Input: Obtain the nucleotide sequences (5' to 3') for all primers to be used in the assay.
- Dimer Analysis: Use oligonucleotide analysis software (e.g., Thermo Fisher's Multiple Prime Analyzer, IDT's OligoAnalyzer) to check for inter-primer homology (complementarity between forward and reverse primers) and intra-primer homology (self-complementarity) [5] [17].
- Parameter Screening:
- Check for complementary regions of 3 or more bases, especially at the 3' ends.
- Calculate the Gibbs free energy (ΔG) of potential dimer formations. More negative ΔG values indicate more stable, and therefore more problematic, dimers.
- Analyze potential hairpin formation within long primers (e.g., LAMP FIP/BIP primers) using tools like mFold [5].
- Iterative Redesign: If stable dimers or hairpins (e.g., ΔG < -5 kcal/mol) are predicted, modify the primer sequences by shifting a few nucleotides upstream or downstream while maintaining a melting temperature (Tₘ) within 65°C–75°C and a GC content between 40%–60% [17].
Protocol: Thermodynamic Evaluation of Dimer Stability
- Apply the Nearest-Neighbor (NN) Model: This model estimates the stability of nucleic acid duplexes based on the identity and orientation of neighboring base pairs. It is the standard for predicting hybridization thermodynamics [5].
- Input Parameters: The model requires the dimer sequence, monovalent salt concentration (e.g., [Na⁺]), and total oligonucleotide strand concentration.
- Calculate ΔG and Tₘ: The software will compute the overall ΔG and Tₘ for the putative dimer. This quantitative data allows for the comparison of different primer sequences and the selection of sets with minimal propensity for interaction [5].
Empirical Validation and Optimization
Theoretical predictions must be confirmed experimentally.
Protocol: Gel Electrophoresis for Dimer Detection
- Run the Amplification Reaction: Perform a no-template control (NTC) reaction containing all components (primers, buffer, polymerase, dNTPs) except the target DNA/RNA.
- Post-Amplification Analysis: Resolve the NTC reaction products on a 2-4% high-resolution agarose or polyacrylamide gel alongside a DNA ladder.
- Visualization: Stain the gel with an intercalating dye like ethidium bromide or SYBR Safe and visualize under UV light.
- Interpretation: The presence of a low molecular weight band (typically smaller than the expected amplicon) in the NTC lane indicates primer dimer formation. The intensity of the band correlates with the efficiency of dimer amplification [3].
Protocol: Real-Time Monitoring with Intercalating Dyes
- Reaction Setup: Set up the NTC reaction in the presence of a fluorescent double-stranded DNA (dsDNA) intercalating dye, such as SYTO 9, SYBR Green, or EvaGreen.
- Real-Time Monitoring: Run the amplification on a real-time PCR instrument, monitoring fluorescence throughout the cycling process.
- Data Interpretation: A slowly rising fluorescent baseline in the NTC, or an amplification curve with a high Cq value, is indicative of non-specific amplification, including primer dimer formation [5]. This is a highly sensitive method for detecting low levels of dimerization.
Table 3: The Scientist's Toolkit: Key Reagents for Studying Hydrogen Bonding and Primer Dimers
| Reagent / Material | Function / Role in Analysis |
|---|---|
| Bst 2.0 WarmStart DNA Polymerase | A common enzyme used in isothermal amplification (e.g., LAMP). Its strand-displacing activity is essential for these techniques [5]. |
| SYTO 9 / SYBR Green Dyes | Fluorescent dsDNA intercalating dyes. Used for real-time monitoring of DNA amplification, allowing detection of both specific and non-specific (e.g., primer dimer) products [5]. |
| AMV Reverse Transcriptase | Used in RT-LAMP to first convert target RNA into complementary DNA (cDNA) before amplification [5]. |
| Betaine | A reagent added to LAMP and PCR buffers to reduce secondary structure in DNA and improve amplification efficiency, especially of GC-rich targets [5]. |
| dNTPs (dATP, dCTP, dGTP, dTTP) | The building blocks (deoxynucleotide triphosphates) used by DNA polymerase to synthesize new DNA strands [5] [3]. |
| MgSO₄ | A source of Magnesium ions (Mg²⁺), which is a essential cofactor for DNA polymerase activity. Its concentration must be optimized, as high levels can promote non-specific priming [5] [3]. |
| Quencher Probes (for QUASR) | Short oligonucleotides with a quencher molecule used in the QUASR detection method. They quench fluorescence of unincorporated labeled primers, reducing background and improving signal-to-noise for specific amplicons [5]. |
Hydrogen bonding provides the specific molecular recognition code that governs G-C and A-T base pairing, a principle that is as fundamental to modern molecular diagnostics as it is to the central dogma of biology. While G-C pairs, with their three hydrogen bonds, confer greater thermodynamic stability than two-bonded A-T pairs, the reality is more complex, with stacking interactions and solvation playing dominant roles in overall duplex stability. In the context of primer design, an over-reliance on simple GC content for predicting stability can be misleading. A thorough thermodynamic analysis, including the evaluation of inter-primer complementarity and secondary structure, is imperative. By applying the quantitative principles and experimental protocols outlined in this guide—from in silico ΔG calculations to empirical validation with NTCs—researchers can systematically design robust assays. Mastering the chemistry of hydrogen bonding enables scientists to harness its power for specificity while mitigating its potential to create artifacts, thereby ensuring the fidelity and reliability of genetic analysis.
Primer-dimer formation represents a significant impediment to the efficiency and specificity of polymerase chain reaction (PCR) assays, a cornerstone of modern molecular biology and diagnostic applications. This whitepaper delineates the fundamental role of hydrogen bonding in the molecular architecture of primer-dimers, with a specific focus on the contributions of guanine (G) and cytosine (C) nucleotide content. The strength of hydrogen bonding, which is markedly greater in G-C pairs (three bonds) than in A-T pairs (two bonds), directly influences the stability of these aberrant primer-primer interactions [18]. We explore how excessive GC content and improperly configured GC clamps at the 3' end of primers can inadvertently promote dimerization, leading to false-positive results and reduced amplification yield. This technical guide provides a detailed examination of the underlying mechanisms, summarizes optimal design parameters in structured tables, and outlines robust experimental protocols for in silico and empirical validation of primer sets to mitigate these issues, thereby enhancing the fidelity of PCR-based research and diagnostics.
The polymerase chain reaction is an indispensable technique across biomedical research, clinical diagnostics, and forensic science. Its success, however, is critically dependent on the specific annealing of oligonucleotide primers to their intended target sequences. A pervasive challenge in PCR optimization is the formation of primer-dimers, which are spurious amplification products generated when primers anneal to each other or to themselves instead of the target DNA template [17] [18]. These artifacts consume reaction reagents, compete with the desired amplification product, and can lead to erroneous interpretations, particularly in sensitive applications like real-time PCR [19].
The formation of primer-dimers is fundamentally governed by the thermodynamics of hydrogen bonding between complementary nucleotide bases. The differential strength between G-C and A-T base pairing is a primary structural culprit in this process. Each G-C pair forms three hydrogen bonds, conferring significantly greater stability to the duplex than an A-T pair, which forms only two [18] [20]. Consequently, primers with high overall GC content or localized regions rich in G and C bases, particularly at the 3' end, possess a heightened propensity for stable, non-specific interactions. This whitepaper examines the precise mechanisms by which GC content and the design of the GC clamp influence dimer formation, framing the discussion within the broader context of hydrogen bonding energetics. The objective is to provide researchers and drug development professionals with a comprehensive framework for designing and validating primers that minimize these detrimental interactions.
Molecular Mechanisms: Hydrogen Bonding and Dimer Energetics
The Biochemistry of Base Pairing
The stability of any nucleic acid duplex, whether a correctly annealed primer-template complex or an erroneous primer-dimer, is predominantly determined by the cumulative strength of hydrogen bonds between opposing bases and the stabilizing effect of base stacking. The hydrogen bond is a key intermolecular force where a hydrogen atom covalently bonded to an electronegative atom (such as nitrogen or oxygen in DNA bases) experiences an attractive force with another electronegative atom. In the context of primer binding, this translates to a direct relationship between base composition and duplex melting temperature (Tm): the temperature at which 50% of the duplex dissociates into single strands [18].
The following dot code illustrates the fundamental relationship between base pairing, hydrogen bonding, and the subsequent risk of primer-dimer formation.
GC Clamp: A Double-Edged Sword
A GC clamp refers to the strategic placement of G or C bases within the last five nucleotides at the 3' end of a primer. The rationale for its use is sound: promoting strong, specific binding at the primer's 3' terminus, which is crucial for enzymatic elongation by DNA polymerase [17] [21]. The stronger hydrogen bonding of a GC clamp helps ensure the primer's 3' end remains securely annealed to the template.
However, this very feature becomes a liability when primers engage in off-target interactions. A strong GC clamp at the 3' end can facilitate the initiation and stabilization of primer-dimer complexes. If the 3' ends of two primers contain complementary sequences with high GC content, the strong hydrogen bonding can effectively "lock" them in place, allowing DNA polymerase to extend them into a dimer product [20]. This is why it is recommended to avoid more than three G or C bases in the last five bases at the 3' end, as this significantly increases the risk of non-specific binding and false-positive results [17] [18] [21].
Quantitative Design Parameters for Optimal Primer Specificity
Adherence to established quantitative parameters during primer design is the most effective strategy for preemptively minimizing dimer formation. The following table consolidates critical design criteria based on widely accepted principles and experimental validations [17] [18] [20].
Table 1: Optimal Primer Design Parameters to Minimize Dimer Formation
| Parameter | Recommended Range | Rationale and Impact on Dimer Formation |
|---|---|---|
| Primer Length | 18-30 nucleotides [17] | Shorter primers (<18 bp) bind more efficiently but may lack specificity; longer primers can increase chances of inter-primer homology. |
| GC Content | 40-60% [17] [18] | Content below 40% results in weak binding; above 60% promotes overly stable non-specific interactions and dimers. |
| GC Clamp (3' end) | 1-3 G/C bases in the last 5 bases [17] [21] | Fewer than 1 reduces binding efficiency; more than 3 G/C bases drastically increases stability of primer-dimers. |
| Melting Temperature (Tm) | 65-75°C for primers in a pair, within 5°C of each other [17] | Ensures both primers anneal efficiently at the same temperature, preventing single-primer artifacts that can lead to dimers. |
| Self-Complementarity | Minimize; avoid runs of 4+ identical bases or dinucleotide repeats (e.g., ACCCC, ATATAT) [17] [22] | Repetitive sequences and mononucleotide runs increase the potential for intra- and inter-primer homology, facilitating dimerization. |
The strategic application of these parameters requires the use of sophisticated bioinformatics tools. These tools perform critical in silico checks for self-complementarity and hairpin formation, parameters that are quantitatively represented as "self-complementarity" and "self 3'-complementarity" scores. The guiding principle for these scores is that lower values are superior, indicating a reduced potential for secondary structure formation [18].
Experimental Protocols for Validation
In Silico Design and Specificity Workflow
Prior to synthesizing primers, a rigorous computational validation workflow is essential. This multi-step process leverages specialized software to preemptively identify and eliminate primers with a high propensity for dimerization [19].
Table 2: Key Research Reagent Solutions for Primer Design and Validation
| Reagent / Tool Category | Specific Examples | Function in Preventing Primer-Dimers |
|---|---|---|
| Primer Design Algorithms | Primer3 (integrated into Primer-BLAST) [23], PrimerMapper [22] | Automates the application of design parameters from Table 1, calculating Tm, GC%, and filtering primers with high self-complementarity. |
| Specificity Checking Tools | NCBI Primer-BLAST [23] | Checks candidate primer pairs for specificity against a selected database (e.g., RefSeq mRNA) to ensure they will not anneal to non-target sequences. |
| Post-Hoc Analysis Tools | URAdime [24] | Analyzes sequencing data from a multiplex PCR to identify the specific primers responsible for generating primer-dimer and super-amplicon artifacts. |
| Dimer Prediction Algorithms | Simulated Annealing Design using Dimer Likelihood Estimation (SADDLE) [24], PrimerMapper's "Multiplex PCR dimer scores" [22] | Systematically calculates cross-complementarity scores between all possible primer pairs in a multiplex set to flag potential dimers before ordering. |
Empirical Optimization and Troubleshooting
Even the most rigorous in silico design requires empirical validation. The following protocol outlines a systematic approach for testing and optimizing primer pairs, with a focus on eliminating spurious amplification [19].
Protocol: Empirical Validation and Optimization of Primer Sets
Reaction Setup: Prepare a standard PCR reaction mix, including all components (polymerase, dNTPs, buffer, Mg²⁺) and the forward and reverse primers. It is critical to include a no-template control (NTC) containing all components except the DNA template. The NTC is essential for detecting primer-dimer formation.
Annealing Temperature Gradient: Perform a thermal cycling reaction using a gradient PCR instrument. Set a range of annealing temperatures (e.g., from 5°C below to 5°C above the calculated Tm of the primers). This helps determine the optimal temperature for specific primer binding.
Primer Concentration Titration: If dimer persistence is observed, titrate the primer concentrations. Testing a range from 50 nM to 500 nM final concentration can identify a concentration that supports efficient amplification while minimizing dimer artifacts. A balanced concentration of both primers is crucial [18].
Analysis: Analyze the PCR products using agarose gel electrophoresis. A successful reaction should show a single, sharp band of the expected amplicon size in the sample lanes, with a clear NTC. A smear or a low molecular weight band (~50 bp or below) in the NTC indicates significant primer-dimer formation.
Troubleshooting: If dimers persist:
- Increase Annealing Temperature: Raise the temperature in 1-2°C increments to disrupt the weaker hydrogen bonding in dimer complexes without significantly affecting specific binding.
- Redesign Primers: If optimization fails, the most reliable solution is to redesign the primers, strictly adhering to the guidelines in Table 1 and avoiding 3' ends with high GC clamps or self-complementary sequences [19].
The intricate role of hydrogen bonding in nucleic acid interactions positions GC content and GC clamp design as critical structural determinants in the formation of primer-dimers. The triple-bonded strength of G-C base pairs, while beneficial for specific target annealing, can readily become a driver of assay failure when primers engage in off-target interactions. A comprehensive strategy that integrates disciplined in silico design, adhering to quantitative parameters for GC distribution and 3' end sequence, with systematic empirical validation is paramount. For the research and drug development community, mastering the hydrogen bonding principles that underpin these artifacts is not merely a technical exercise but a prerequisite for achieving the robust, reliable, and reproducible results demanded in modern molecular biology. As PCR continues to evolve and be applied in increasingly complex multiplexed and diagnostic formats, a deep understanding of these structural culprits will remain essential.
Within the context of primer dimer research, the formation of non-specific amplification artifacts is a critical challenge. These dimers are primarily stabilized by inter-primer hydrogen bonds. This whitepaper examines the underappreciated role of water molecules as direct competitors in these hydrogen bond interactions. The thermodynamic stability of a primer dimer is not merely a function of primer-primer affinity but is a result of a competitive equilibrium between primer-primer, primer-water, and water-water hydrogen bonds. Understanding this solvation shell competition is essential for optimizing assay specificity in molecular diagnostics and drug development targeting nucleic acid interactions.
Quantitative Data on Hydrogen Bond Energetics
The stability of hydrogen-bonded complexes in aqueous solution is governed by the net free energy change, which accounts for the competition between bond formation and the associated (de)solvation penalties.
Table 1: Energetic Contributions to Hydrogen Bond Formation in Aqueous Solution
| Interaction Type | Enthalpy (ΔH, kJ/mol) | Entropy (TΔS, kJ/mol) | Free Energy (ΔG, kJ/mol) | Context in Primer Dimers |
|---|---|---|---|---|
| Base Pair (e.g., A-T) | -15 to -25 (Favorable) | -10 to -20 (Unfavorable) | -4 to -8 (Net Favorable) | Direct stabilization of the dimer complex. |
| Water-Water (Bulk) | ~ -20 (Favorable) | + (Favorable) | ~ -10 (Net Favorable) | Represents the stable reference state for displaced water. |
| Polar Group Hydration | -20 to -40 (Favorable) | -15 to -30 (Unfavorable) | -5 to -10 (Net Favorable) | Energy cost to dehydrate primer bases before dimer formation. |
| Net Dimer Formation | ~0 to -10 (Slightly Favorable) | -15 to -30 (Highly Unfavorable) | +5 to +20 (Net Unfavorable) | Overall process including dehydration and base pairing. |
Experimental Protocols for Probing Water Competition
3.1. Isothermal Titration Calorimetry (ITC) for Thermodynamic Profiling
Objective: To directly measure the enthalpy (ΔH), stoichiometry (N), and equilibrium constant (Ka) of primer-primer binding, thereby deriving the full thermodynamic profile (ΔG, ΔS).
Protocol:
- Sample Preparation: Dissolve the forward and reverse primers in an identical, degassed buffer (e.g., 10 mM Sodium Phosphate, pH 7.0, 100 mM NaCl). The concentration of the primer in the syringe (typically 100-200 µM) should be 10-20 times higher than the concentration in the cell (typically 10 µM).
- Instrument Setup: Thoroughly clean and dry the ITC sample cell and syringe. Load the reverse primer solution into the 1.4 mL sample cell and the forward primer into the 250 µL syringe. Set the reference cell to degassed, pure water.
- Titration Parameters:
- Temperature: 25°C
- Number of Injections: 19
- Injection Volume: 2 µL (first injection of 0.4 µL discarded from data analysis)
- Duration: 4 seconds per injection
- Spacing: 180 seconds between injections
- Stirring Speed: 750 rpm
- Control Experiment: Perform an identical titration of the forward primer into buffer alone to measure and subtract the heat of dilution.
- Data Analysis: Fit the corrected isotherm (heat per mole of injectant vs. molar ratio) using a suitable binding model (e.g., "One Set of Sites") to obtain ΔH, Ka, and N. Calculate ΔG using ΔG = -RT lnKa and ΔS using ΔG = ΔH - TΔS.
3.2. Molecular Dynamics (MD) Simulations with Explicit Solvent
Objective: To visualize and quantify the dynamics of water molecules in the solvation shell of primers and during dimer formation.
Protocol:
- System Setup:
- Construct a model of a primer dimer using nucleic acid building blocks or extract a structure from a relevant PDB file.
- Place the dimer in a simulation box (e.g., a rhombic dodecahedron) with a minimum 1.0 nm distance between the solute and the box edge.
- Solvate the system with explicit water models (e.g., TIP3P, SPC/E).
- Add ions (e.g., Na+, Cl-) to neutralize the system and achieve a physiologically relevant salt concentration (e.g., 150 mM NaCl).
- Energy Minimization: Perform steepest descent energy minimization to remove steric clashes and bad contacts.
- Equilibration:
- Run a 100 ps simulation in the NVT ensemble (constant Number of particles, Volume, and Temperature) using a thermostat (e.g., V-rescale) to stabilize the temperature at 300 K.
- Follow with a 100 ps simulation in the NPT ensemble (constant Number of particles, Pressure, and Temperature) using a barostat (e.g., Berendsen) to stabilize the pressure at 1 bar.
- Production Run: Execute a long-term MD simulation (e.g., 100-500 ns) in the NPT ensemble, saving atomic coordinates every 10 ps.
- Trajectory Analysis:
- Hydrogen Bond Analysis: Use tools like
gmx hbond(GROMACS) to calculate the lifetime and occupancy of hydrogen bonds between primers and between primers and water. - Radial Distribution Function (RDF): Calculate g(r) between primer atoms and water oxygen to understand solvation structure.
- Energetics: Decompose interaction energies to quantify the contribution of water displacement to dimer stability.
- Hydrogen Bond Analysis: Use tools like
Visualizations
Title: Water Competition in Dimer Formation
Title: ITC Experimental Workflow
The Scientist's Toolkit
Table 2: Essential Research Reagents and Materials
| Item | Function/Benefit |
|---|---|
| Ultra-Pure DNase/RNase-Free Water | Eliminates nuclease contamination and ensures a consistent, pure aqueous environment for studying hydrogen bonding. |
| Isothermal Titration Calorimeter (ITC) | The gold-standard for label-free, in-solution measurement of binding thermodynamics, directly quantifying the heat changes from competitive interactions. |
| Molecular Dynamics Software (e.g., GROMACS, AMBER) | Enables atomistic simulation of primer-water and primer-primer interactions with explicit solvent models over time. |
| Explicit Solvent Force Fields (e.g., OPC, TIP4P-Ew) | Advanced water models that provide a more accurate representation of hydrogen bond geometry and energetics compared to simpler models. |
| Controlled Atmosphere Glove Box | Allows for the preparation of samples in a water vapor-free environment (e.g., using N₂ gas) to study the effects of controlled re-hydration. |
| High-Performance Salt Solutions (e.g., NaCl, MgCl₂) | Used to systematically investigate the ionic strength's effect on water structure and its competition for phosphate backbone interactions. |
In molecular biology, the efficacy of polymerase chain reaction (PCR) and related amplification techniques is fundamentally dependent on the precise binding of primers to their target DNA sequences. The formation of primer secondary structures, particularly hairpins, represents a significant challenge to assay performance. These structures are stabilized primarily by intramolecular hydrogen bonding, which can sequester primer sequences into inactive conformations [5]. When primers form stable hairpins, they are unable to anneal to the template DNA, leading to reduced amplification efficiency, false negatives, or non-specific amplification [25] [4]. Within the broader context of hydrogen bonding research in primer-dimers, understanding hairpin formation is paramount, as these intramolecular interactions follow the same thermodynamic principles that govern intermolecular primer-dimer artifacts.
The propensity for hairpin formation is intrinsically linked to the molecular composition of the primer. Hydrogen bonds between complementary base pairs within a single oligonucleotide strand facilitate the folding of the molecule onto itself. Guanine-cytosine (G-C) base pairs, connected by three hydrogen bonds, confer greater stability to these secondary structures than adenine-thymine (A-T) pairs, which are connected by only two hydrogen bonds [18]. Consequently, primers with high GC content, especially in self-complementary regions, are particularly prone to forming stable hairpins that can withstand the annealing temperature of a PCR reaction, thereby compromising the experiment's success [4].
Molecular Mechanisms of Hairpin Formation and Stabilization
Thermodynamic Principles and Hydrogen Bonding
Hairpin formation is a spontaneous process driven by a negative change in Gibbs free energy (ΔG), which indicates the stability of the formed structure [4]. The overall stability of a hairpin is determined by the sum of favorable and unfavorable energy contributions. The favorable energy gain comes from the hydrogen bonds formed between complementary bases and the stacking interactions between adjacent base pairs in the stem region. Conversely, the main unfavorable energy component is the loop entropy, which is required to bring the complementary regions together to form the stem [5]. The nearest-neighbor model is widely used to predict the thermodynamic stability of these secondary structures by considering the sequence context and interactions between adjacent nucleotide pairs [5].
The role of intramolecular hydrogen bonding in shaping molecular conformation extends beyond nucleic acids and is a critical factor in drug design and bioavailability. Research on small drug molecules like piracetam has demonstrated that the formation of intramolecular hydrogen bonds (IMHBs) can significantly alter a compound's properties, facilitating passive diffusion across lipid membranes by reducing the polarity and desolvation penalty [26]. This principle of conformational control via internal hydrogen bonding is analogous to its role in primer biochemistry, where IMHBs dictate the folding and functional availability of the oligonucleotide.
Structural Types and Classification
Hairpin structures are primarily categorized based on the location of the self-complementary region, which determines their potential impact on amplification:
- 3' End Hairpins: Formed when the first and last few bases at the 3' end of a primer are complementary. These are the most detrimental to PCR efficiency because they can prevent the DNA polymerase from binding and initiating extension [4]. A stable 3' end hairpin effectively blocks the primer from functioning.
- Internal Hairpins: Occur when complementary sequences are located within the internal regions of the primer. While generally less catastrophic than 3' end hairpins, stable internal structures can still sequester a significant portion of the primer molecule, reducing the effective concentration of available primers and lowering the reaction yield [4].
Table 1: Characteristics and Impacts of Different Hairpin Types.
| Hairpin Type | Structural Feature | Potential Impact on PCR |
|---|---|---|
| 3' End Hairpin | Complementarity between the first and last 3-4 bases at the 3' end. | Prevents polymerase binding and extension; most severe impact. |
| Internal Hairpin | Complementarity between two internal regions, creating a loop. | Reduces primer availability and efficiency; can cause failure. |
| Stable Hairpin | ΔG value more negative than -3 kcal/mol [4]. | May not denature at PCR annealing temperature. |
| Unstable Hairpin | ΔG value less negative than -3 kcal/mol [4]. | Likely to denature during PCR, minimal impact. |
Experimental Detection and Analysis Methodologies
In Silico Prediction and Analysis Workflow
Computational tools are the first line of defense against hairpin formation in primer design. The following workflow outlines a standard protocol for analyzing potential secondary structures.
Title: In Silico Primer Analysis Workflow
Protocol Steps:
- Sequence Input and Hairpin Prediction: Input the candidate primer sequence (typically 18-24 nucleotides) into a dedicated analysis tool such as IDT's OligoAnalyzer or mFold [5] [4]. These programs simulate the folding of the single-stranded DNA and identify regions with intra-primer homology capable of forming hairpin loops.
- Thermodynamic Stability Assessment: The software calculates the Gibbs free energy (ΔG) for all possible secondary structures using the nearest-neighbor model [5]. As a general rule, hairpins with a ΔG value more negative than -3 kcal/mol for internal hairpins or -2 kcal/mol for 3' end hairpins are considered stable and likely to interfere with the PCR reaction [4].
- 3' End Complementarity Check: A critical step is to manually inspect the prediction results for any complementarity at the 3' terminus. Even one or two complementary bases at the 3' end can form a self-amplifying structure, leading to primer extension and significant non-specific background amplification [5].
- Cross-homology and Dimer Analysis: Use tools like the Thermo Fisher Scientific Multiple Primer Analyzer to check for inter-primer homology that could lead to primer-dimer formation between forward and reverse primers [27] [4]. Furthermore, test primer specificity by performing a BLAST search against the relevant genomic database to ensure the primer binds uniquely to the intended target [25] [4].
Empirical Validation Protocols
While in silico analysis is powerful, empirical validation is essential for confirming primer performance in actual reaction conditions.
Protocol: Gradient PCR with Melt Curve Analysis
Reaction Setup:
- Prepare a standard PCR master mix containing buffer, dNTPs, DNA polymerase (e.g., Bst 2.0 WarmStart for LAMP), and a fluorescent intercalating dye like SYTO 9 or SYBR Green [5] [4].
- Add the forward and reverse primers at their working concentrations (typically 0.1-0.5 µM each for standard PCR) [5].
- Include a no-template control (NTC) to detect amplification arising from primer artifacts alone.
Thermal Cycling:
- Perform a temperature gradient PCR, setting the annealing temperature (Ta) to a range from 5°C below to 5°C above the calculated theoretical Ta [25] [4].
- Following amplification, conduct a high-resolution melt curve analysis by gradually increasing the temperature from 60°C to 95°C while continuously monitoring fluorescence.
Data Interpretation:
- Analyze the amplification curves. A slowly rising baseline in the NTC, or early amplification in the NTC, indicates non-specific amplification potentially due to hairpins or primer-dimers [5].
- Examine the melt curve. A single, sharp peak corresponds to the specific PCR product. Multiple or broad peaks suggest the presence of non-specific products or primer artifacts, which may stem from primers with stable secondary structures [4].
Quantitative Data and Design Parameters
Adherence to established primer design parameters is the most effective strategy to minimize the risk of hairpin formation. The following table summarizes the optimal values for key design characteristics based on current research and best practices.
Table 2: Optimal Primer Design Parameters to Minimize Secondary Structures [25] [4] [18].
| Design Parameter | Optimal Value or Range | Rationale and Impact on Hairpins |
|---|---|---|
| Primer Length | 18 - 24 bp | Balances specificity and binding efficiency; overly long primers (>30 bp) increase the chance of intra-primer homology. |
| Melting Temperature (Tm) | 50 - 65 °C; within 5 °C for a pair | Ensures both primers anneal simultaneously. A very low Tm can permit hairpin stability during annealing. |
| GC Content | 40 - 60% | Provides sufficient stability without promoting overly stable G-C rich hairpins (G-C bonds have 3 H-bonds vs. A-T's 2). |
| GC Clamp | 2-3 G/C bases in last 5 bases at 3' end | Stabilizes primer-template binding but more than 3 can cause non-specific binding and increase risk of 3' end hairpins. |
| Self-Complementarity | As low as possible | Directly measures the potential for a primer to form hairpins or self-dimers. |
| ΔG of Hairpins | > -3 kcal/mol (internal), > -2 kcal/mol (3' end) | A less negative ΔG ensures the hairpin is unstable and will denature at the reaction temperature. |
Case Study: Troubleshooting Hairpins in Complex Assays
The impact of hairpins is particularly pronounced in techniques involving multiple long primers, such as Loop-Mediated Isothermal Amplification (LAMP). A study on RT-LAMP detection of dengue and yellow fever viruses provides a compelling case study [5].
Experimental Observation: Previously published primer sets displayed a slowly rising baseline in real-time fluorescence curves and poor endpoint signal in the QUASR detection technique. This was hypothesized to be due to amplifiable primer dimers and self-amplifying hairpin structures in the FIP and BIP primers, which are typically 40-45 bases long [5].
Methodology and Intervention:
- The researchers performed a thorough thermodynamic analysis of the original primers using the nearest-neighbor model to identify regions of high self-complementarity.
- Minor sequence modifications were introduced to these regions to disrupt the stable secondary structures, particularly those with 3' complementarity, while preserving target specificity.
- The performance of the original and modified primer sets was compared using real-time RT-LAMP with intercalating dyes and the QUASR endpoint technique.
Results: The modified primers, designed to eliminate amplifiable hairpins, demonstrated significantly improved performance. The non-specific background amplification was dramatically reduced, leading to clearer positive-negative discrimination and more reliable assay outcomes [5]. This study quantitatively demonstrates that even hairpins with complementarity one or two bases away from the 3' end can self-amplify and that their elimination is critical for robust assay performance.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Analyzing Primer Secondary Structures.
| Reagent / Tool | Function and Application |
|---|---|
| Bst 2.0 WarmStart DNA Polymerase | An enzyme commonly used in LAMP assays. Its strand-displacing activity is sensitive to primer secondary structures, making it a good tool for testing functional primer performance [5]. |
| SYTO 9 / SYBR Green Dyes | Fluorescent intercalating dyes that bind double-stranded DNA. They are used in real-time PCR/LAMP to monitor amplification kinetics and identify non-specific amplification from primer artifacts [5] [4]. |
| IDT OligoAnalyzer Tool Suite | A web-based suite for in silico primer analysis. It calculates Tm, ΔG for hairpins and self-dimers, and visualizes potential secondary structures [5] [4]. |
| Thermo Fisher Multiple Primer Analyzer | A tool for checking cross-dimer formation between forward and reverse primers, as well as self-dimerization, which often co-occurs with hairpin problems [27] [4]. |
| mFold Software | A tool for predicting the secondary structure formation of nucleic acids, used for advanced folding simulations and stability assessments [5]. |
The formation of hairpin structures through intramolecular hydrogen bonding is a fundamental phenomenon that can critically undermine the success of nucleic acid amplification experiments. The thermodynamic principles that govern these interactions are well-characterized, enabling robust in silico prediction methods. By integrating computational design adhering to strict parameters, such as optimizing length, Tm, and GC content, with empirical validation techniques like gradient PCR, researchers can effectively mitigate the risks posed by primer secondary structures. As demonstrated in advanced applications like LAMP, a meticulous approach to primer design—paying particular attention to 3' end stability—is not merely a preliminary step but a central component in the development of specific, sensitive, and reliable molecular assays. Future research into the dynamics of intramolecular hydrogen bonding will continue to refine our understanding and control of these crucial molecular interactions.
From Theory to Bench: Strategic Primer Design and Dimer Detection Methods
The polymerase chain reaction (PCR) stands as one of the most pivotal inventions in molecular biology, enabling the amplification of small amounts of genetic material for identification, manipulation, and detection [28]. At the heart of every successful PCR experiment lies a critical component: well-designed primers. The quality of these primers directly governs the specificity, efficiency, and overall success of the amplification reaction [29]. In the context of drug development and advanced research, where reproducibility and accuracy are paramount, adhering to gold-standard design rules transcends mere recommendation and becomes an absolute necessity. The exquisite specificity and sensitivity that make PCR uniquely powerful are controlled predominantly by primer properties [30]. Consequently, poor design combined with failure to optimize reaction conditions frequently results in reduced technical precision, false positives, or false negative detection of amplification targets.
The foundational principles of primer design extend beyond simple sequence selection to encompass a deep understanding of molecular interactions, particularly hydrogen bonding dynamics. These interactions not only facilitate the specific binding of primers to their target sequences but also govern undesirable side reactions such as primer-dimer formation. When primers interact with each other instead of the target template, they form primer-dimers through hydrogen bonding between complementary bases, effectively competing for precious reaction resources and compromising assay sensitivity [31] [32]. This comprehensive guide details the gold-standard parameters for primer design, with particular emphasis on how the rules governing length, melting temperature (Tm), and GC content directly influence hydrogen bonding stability and specificity, ultimately determining experimental outcomes in research and diagnostic applications.
Core Parameters for Gold-Standard Primer Design
Optimal Primer Length
Primer length fundamentally determines the balance between specificity and binding efficiency. Excessively short primers lack the sequence complexity required for unique targeting, while overly long primers exhibit slower hybridization rates and can create unnecessarily high melting temperatures that hinder polymerase function.
- Optimal Range: The consensus across multiple authoritative sources recommends maintaining primer lengths between 18 and 30 nucleotides [31] [17] [33]. The most frequently utilized and recommended range narrows this to 18–24 bases [18] [28]. This length provides sufficient sequence for unique binding within complex genomes while ensuring efficient annealing during thermal cycling.
- Impact of Deviations: Primers shorter than 18 bases risk insufficient specificity and may anneal to multiple non-target sites, leading to nonspecific amplification [29]. Conversely, primers exceeding 30 bases hybridize more slowly to their target sequence, potentially reducing amplification yield and efficiency [18]. Long primers also tend to form more stable secondary structures, further complicating the reaction dynamics.
Melting Temperature (Tm)
The melting temperature (Tm) of a primer is defined as the temperature at which half of the DNA duplex dissociates into single strands, providing a quantitative measure of duplex stability [28]. Accurate Tm calculation and synchronization between primer pairs is arguably the most critical factor in successful PCR design.
- Optimal T
mRange: While specific recommended values vary slightly, the general consensus falls within a 55–65°C range [34] [31]. For standard PCR, primers with Tmvalues of 52–58°C often produce excellent results, while for qPCR applications, an optimal Tmof 60–64°C is frequently advised [33] [28]. - Primer Pair Matching: Perhaps the most critical rule is that the forward and reverse primers in a pair should have closely matched T
mvalues, ideally within ≤2°C of each other [33] [18]. A mismatch of 5°C or more can lead to failed amplification, as one primer will anneal efficiently while the other does not [28]. - Calculation Methods: Two primary methods exist for T
mcalculation:- Basic GC% Formula: T
m= 4(G + C) + 2(A + T). This method provides a rough estimate but is less accurate as it ignores sequence context [18]. - Nearest-Neighbor Thermodynamics: This is the gold-standard method based on SantaLucia's unified parameters (1998). It accounts for dinucleotide stacking energies and sequence context, achieving prediction accuracy of ±1–2°C [34]. It requires specialized software but is incorporated into all modern primer design tools.
- Basic GC% Formula: T
Table 1: Comparison of Melting Temperature Calculation Methods
| Method | Accuracy | Complexity | Key Consideration |
|---|---|---|---|
| Nearest-Neighbor Thermodynamics | Highest (±1–2°C) [34] | High | Accounts for dinucleotide stacking, sequence context, and salt effects; the gold standard for design [34]. |
| GC% Approximation | Low (±5–10°C) [34] | Low | Considers only GC content, ignoring sequence context; suitable for quick estimates only [34]. |
| Salt-Adjusted Formulas | High | Medium | Incorporates Owczarzy et al. (2008) corrections for mixed ion solutions, essential for PCR with Mg²⁺ [34]. |
GC Content and GC Clamp
The GC content—the percentage of guanine (G) and cytosine (C) bases in the primer—directly influences binding stability through hydrogen bonding. GC base pairs form three hydrogen bonds, while AT pairs form only two [18]. This differential bonding energy is the physical basis for several design rules.
- Optimal GC Content: Primers should have a GC content between 40% and 60%, with an ideal target of 50% [34] [33] [29]. This range provides stable primer-template binding without promoting mispriming or secondary structure formation.
- Consequences of Extreme GC Content:
- Low GC (<40%): Results in overly weak priming sites due to insufficient hydrogen bonding, leading to unstable hybrids and potentially low product yield [34].
- High GC (>60%): Promotes stable non-specific binding and increases the risk of secondary structures (e.g., hairpins) because of the stronger hydrogen bonding, which can halt the reaction [34] [29].
- The GC Clamp: The presence of G or C bases within the last five bases at the 3' end of the primer is known as a GC clamp. This promotes stronger binding at the 3' terminus due to the extra hydrogen bonds, ensuring the polymerase extension site is securely anchored [17] [28]. However, more than three G or C bases in the last five should be avoided, as this can encourage non-specific initiation [28].
Advanced Considerations and Troubleshooting
Avoiding Secondary Structures and Primer Dimers
The same hydrogen bonding forces that facilitate specific primer-template annealing can also lead to detrimental intra-primer and inter-primer interactions. Preventing these is crucial for assay efficiency.
- Hairpins: Formed by intramolecular hybridization within a single primer, where two regions of three or more nucleotides complement each other [18] [28]. Hairpins, especially those at the 3' end, can prevent the primer from binding to its template. Stability is measured by Gibbs Free Energy (ΔG); larger negative values indicate more stable, problematic structures. Hairpins with a ΔG < -3 kcal/mol should generally be avoided [34] [28].
- Self-Dimers and Cross-Dimers: Self-dimers occur when two copies of the same primer hybridize, while cross-dimers form between the forward and reverse primers [18]. These interactions consume primers and polymerase resources, drastically reducing the yield of the desired product. The ΔG of any dimer should be weaker (more positive) than -9.0 kcal/mol [33], with 3' end dimers more critical than internal ones.
Table 2: Summary of Critical Parameters to Avoid in Primer Sequences
| Parameter | Description | Maximum Tolerable Threshold | Impact of Violation |
|---|---|---|---|
| Runs (Homopolymers) | Consecutive identical bases (e.g., AAAA or GGGG) [17]. | 4 contiguous bases [17] [28]. | Mispriming due to slippage, non-specific binding. |
| Dinucleotide Repeats | Short, tandem repeats (e.g., ATATAT) [17]. | 4 di-nucleotides [28]. | Mispriming, poor specificity. |
| Self 3'-Complementarity | Complementarity at the 3' end leading to hairpins [18]. | ΔG > -2 kcal/mol [28]. | Failure of primer extension, no product. |
| Inter-Primer 3'-Complementarity | Complementarity between the 3' ends of forward and reverse primers [31]. | ≤3 contiguous bases, especially at 3'-ends [31]. | Primer-dimer formation, reduced target yield. |
| Intra-Primer Homology | Self-complementarity within a single primer [17]. | ≤3 contiguous bases [31]. | Hairpin formation. |
The Critical Role of the 3' End
The 3' terminus of the primer is where DNA synthesis initiates, making its configuration and stability paramount. A lower ΔG (less negative) at the 3' end is desirable as it facilitates specific binding and reduces the likelihood of non-specific initiation from mismatched primers [29] [28]. Some advanced strategies involve covalently modifying the 3' end with stable alkyl groups attached to the exocyclic amines of adenine or cytosine. These bulky groups are believed to poorly extend from misprimed structures, thereby enhancing specificity by chemically suppressing primer-dimer propagation [32].
Experimental Condition Considerations
Theoretical primer design must be translated into practical reaction environments, which significantly influence hybridization behavior.
- Salt Concentrations: Cations shield the negative charges on the DNA phosphate backbone. Both monovalent (Na⁺, K⁺) and divalent (Mg²⁺) ions stabilize the duplex, increasing the observed T
m.- Mg²⁺ has a stronger effect than Na⁺ and is a critical cofactor for polymerase activity. However, it also binds to dNTPs, reducing its effective concentration. The Owczarzy (2008) salt correction formula is recommended for accurate T
mprediction under standard PCR conditions (1.5–2.5 mM Mg²⁺) [34]. - Impact: Doubling Na⁺ from 50 mM to 100 mM can increase T
mby ~5°C, while adding 2 mM Mg²⁺ can boost Tmby 5–8°C [34].
- Mg²⁺ has a stronger effect than Na⁺ and is a critical cofactor for polymerase activity. However, it also binds to dNTPs, reducing its effective concentration. The Owczarzy (2008) salt correction formula is recommended for accurate T
- Annealing Temperature (T
a) is critically derived from the primer Tm. A common and robust formula is the Rychlik method: TaOpt = 0.3 x Tm(primer) + 0.7 x Tm(product) – 14.9, where Tm(primer) is for the less stable primer [28]. A good starting point is to set Ta2–5°C below the calculated Tmof the primers [18]. If Tais too low, non-specific amplification occurs; if too high, product yield plummets [33]. - Additives: DMSO is often added to disrupt secondary structures, particularly in GC-rich templates. It lowers the T
mby approximately 0.5–0.7°C per 1% concentration and should be accounted for in calculations [34].
Essential Tools and Verification Workflow
The Scientist's Toolkit: Research Reagent Solutions
Modern primer design and validation rely on a suite of sophisticated software tools and reagents.
Table 3: Essential Research Reagents and Tools for Primer Design and Analysis
| Tool/Reagent Category | Specific Examples | Primary Function |
|---|---|---|
| Primer Design Software | Primer3 [31], Primer-BLAST [23], Primer Express (Applied Biosystems) [31], Primer Premier [28] | Automates primer design based on user-defined parameters and template sequence. |
| Oligo Analysis Tools | OligoAnalyzer Tool (IDT) [33], UNAFold Tool [33], Netprimer [31] | Analyzes Tm, secondary structures (hairpins, dimers), and ΔG values for pre-designed oligos. |
| Specificity Check Tools | NCBI BLAST [33] [29], Primer-BLAST [23] | Verifies primer uniqueness against genomic databases to ensure target-specific binding. |
| Hot-Start Polymerases | Antibody-bound or chemically modified Taq polymerases [32] | Reduces non-specific amplification and primer-dimer formation during reaction setup by inhibiting polymerase activity at low temperatures. |
| Chemically Modified Primers | 3'-end alkyl-modified primers [32], 2'-O-methyl RNA, LNA residues [32] | Enhances PCR specificity by sterically hindering the extension of mis-annealed primers. |
Primer Design and Validation Workflow
The following diagram visualizes the systematic, gold-standard workflow for designing and validating PCR primers, integrating both in silico and experimental steps.
Diagram Title: Primer Design and Validation Workflow
A critical final step in the design process is specificity verification. Before synthesizing primers, their sequences must be checked using a BLAST search against the appropriate genomic database (e.g., Refseq mRNA) to ensure they are unique to the intended target [23] [29]. This in silico step prevents costly experimental failures due to off-target amplification.
Adherence to the gold-standard rules of primer design—length (18–30 bp), melting temperature (55–65°C with ≤2°C difference between pairs), and GC content (40–60%)—is non-negotiable for robust, reproducible PCR in research and drug development. These parameters are not arbitrary but are fundamentally rooted in the thermodynamics of hydrogen bonding, which governs the stability of the primer-template duplex and the propensity for aberrant structures like primer-dimers. By meticulously applying these guidelines, leveraging modern design and analysis software, and validating designs experimentally, researchers can ensure that their assays achieve the highest levels of specificity and efficiency, thereby generating reliable and meaningful scientific data.
Leveraging Computational Tools for Predicting Primer-Primer Interactions
In polymerase chain reaction (PCR) experiments, the undesired formation of primer-dimers through hydrogen bonding between primers represents a significant challenge, often leading to reduced amplification efficiency and false results. Primer-dimers are formed due to the presence of complementary sequences within a single primer or between forward and reverse primers, leading to inter-primer homology that enables hydrogen bonding between them [18]. This comprehensive technical guide explores the sophisticated computational tools and methodologies developed to predict and prevent these detrimental interactions, with particular focus on the fundamental role of hydrogen bonding in stabilizing these non-productive complexes. By framing this discussion within the context of molecular interactions, we provide researchers with a detailed roadmap for leveraging computational predictions to enhance experimental outcomes, ultimately improving the reliability of diagnostic assays, research applications, and drug development processes.
The Molecular Basis of Primer-Dimer Formation
Hydrogen Bonding in Nucleic Acid Interactions
The formation of primer-dimers is fundamentally governed by hydrogen bonding between complementary nucleotide bases, with GC base pairs forming three hydrogen bonds and AT base pairs forming two hydrogen bonds [18] [4]. This differential bonding strength directly influences the stability of primer-dimers, with GC-rich regions contributing disproportionately to dimer stability due to their additional hydrogen bond. The Gibbs free energy (ΔG) of these interactions quantifies the spontaneity of dimer formation, with more negative ΔG values indicating stronger, more stable interactions that are more likely to interfere with PCR amplification [4].
When primers fold back on themselves or bind to each other instead of the target template, they create secondary structures that fall into two primary categories [4]:
- Intra-primer homology: A region of 3 or more bases that are complementary to another region within the same primer, causing intramolecular bonding and hairpin formation.
- Inter-primer homology: Forward and reverse primers that have complementary sequences, causing intermolecular bonding and primer-dimer formation.
These interactions are particularly problematic when they occur near the 3' end of primers, as this positioning can lead to extension by DNA polymerase, effectively amplifying the dimerized primers themselves rather than the target template [4].
Table 1: Types of Primer Secondary Structures and Their Characteristics
| Structure Type | Formation Mechanism | ΔG Tolerance (kcal/mol) | Primary Impact |
|---|---|---|---|
| Hairpins | Intra-primer homology; primer folds on itself | > -2 (3' end), > -3 (internal) | Prevents binding to template; reduces efficiency |
| Self-Dimers | Inter-primer homology between identical primers | > -5.0 | Reduces primer availability; competes with target |
| Cross-Dimers | Inter-primer homology between forward and reverse primers | > -5.0 | Creates alternative amplification products; reduces yield |
Experimental Evidence of Hydrogen Bonding Effects
Recent research has quantitatively demonstrated how hydrogen bonding directly impacts primer-template interactions. A 2024 systematic study examining primer-template mismatches revealed that mismatch location significantly influences amplification efficiency, with 3' end mismatches having the most detrimental effects [35]. Furthermore, the study demonstrated that in complex template systems, such as those encountered in metagenomic analysis, mismatch amplifications can dominate despite the presence of perfect match possibilities, highlighting the critical importance of comprehensive computational prediction that accounts for these non-ideal interactions [35].
Advanced analytical techniques, including solid-state NMR spectroscopy relaxation dispersion experiments, have enabled direct measurement of hydrogen bond dynamics in molecular systems [36]. While these studies have primarily focused on telechelic polymers with hydrogen-bonded end groups, the methodologies demonstrate the potential for directly probing the kinetics of molecular-level scission-reaggregation events in hydrogen-bonded systems, offering a potential pathway for future direct investigation of primer-dimer stability [36].
Computational Tools for Predicting Primer-Primer Interactions
Established Primer Design Software
Traditional primer design tools incorporate algorithms to evaluate potential primer-primer interactions, with Primer3 emerging as a community standard for accessible primer design [37] [38]. Primer3 provides basic checks for self-complementarity and self 3'-complementarity, which serve as proxies for predicting dimer formation potential [18]. However, these tools often rely on simplified thermodynamic models and may not comprehensively evaluate all possible interaction configurations, particularly in complex experimental setups involving multiple primer pairs.
For large-scale primer design applications, integrated pipelines like CREPE (CREate Primers and Evaluate) have been developed, which combine the functionality of Primer3 with in-silico PCR (ISPCR) for specificity analysis [37]. This integrated approach performs primer design and specificity analysis through a custom evaluation script that can process any given number of target sites at scale, providing a final output that summarizes the lead primer pair for each target site along with a measure of the likelihood of binding to off-targets [37].
Table 2: Computational Tools for Primer Design and Evaluation
| Tool Name | Primary Function | Interaction Prediction Method | Scale Capability |
|---|---|---|---|
| Primer3 | Basic primer design | Self-complementarity checks | Single targets |
| CREPE | Large-scale design with evaluation | Primer3 + ISPCR specificity analysis | Hundreds of targets |
| RNN-based Prediction | PCR success prediction | Machine learning from sequence relationships | Limited only by training data |
| Benchling | Comprehensive primer analysis | ΔG calculations for secondary structures | Single targets with visualization |
Advanced Machine Learning Approaches
Recent advances in machine learning have introduced novel methods for predicting PCR success based on primer and template sequences. A 2021 study developed a recurrent neural network (RNN) approach that expresses primer-template interactions as five-lettered "pseudo-sentences" to predict amplification success with approximately 70% accuracy [39]. This method comprehensively evaluates various relationships, including hairpin structures, primer dimers, and partial complementarities, by converting these interactions into symbolic representations that can be processed by natural language processing algorithms [39].
The RNN model was trained on experimental data from 72 primer sets tested against 31 DNA templates, with PCR results serving as ground truth for supervised learning [39]. This approach demonstrates the potential for machine learning models to capture complex relationships that may be missed by traditional thermodynamic calculations alone, particularly when predicting "non-amplifying" combinations that are crucial for avoiding false positives in diagnostic applications [39].
Specificity Validation Tools
After initial primer design, validation of specificity is essential to minimize off-target effects. The Basic Local Alignment Search Tool (BLAST) is widely used to identify regions of significant cross-homology by comparing primer sequences against genetic databases [38]. This process helps ensure that primers are specific to the intended target sequence and will not anneal to unrelated regions in the genome [38].
For more specialized applications, tools such as In-Silico PCR (ISPCR) can be deployed from the command line to identify potential off-target binding sites, including those with imperfect matches that might still result in aberrant PCR products [37]. Advanced implementations allow for parameter adjustments to control the stringency of these searches, enabling researchers to balance specificity with practical amplification constraints [37].
Experimental Protocols for Validation
Deconstructed PCR for Empirical Measurement
The Deconstructed PCR (DePCR) methodology provides a quantitative experimental system for interrogating primer-template interactions by separating linear copying of templates from exponential amplification [35]. This approach preserves information about which primers anneal to source DNA templates—information that is typically lost in standard PCR due to the "scrambling" of primer-template interactions over multiple cycles [35].
Protocol: Deconstructed PCR for Primer Interaction Analysis
Template Preparation: Synthesize double-stranded DNA templates with unique priming sites and recognition sequences. For systematic studies, introduce controlled variations at specific positions (e.g., -2, -8, and -14 bases from the 3' end) [35].
Primer Design: Synthesize primers with varying degrees of mismatch (0, 1, 2, or 3 mismatches) at defined positions relative to the template sequences [35].
Linear Copying Phase: Perform initial cycles with separation of primer annealing to source DNA from amplification. This preserves the identity of primers annealing to original templates [35].
Exponential Amplification: Begin standard PCR amplification using the products from the linear copying phase.
Sequencing and Analysis: Sequence amplification products using high-throughput platforms (e.g., Illumina MiniSeq) and quantify amplicon representation to determine primer binding efficiency [35].
This protocol enables direct measurement of primer-template interactions and has demonstrated that heavily degenerate primer pools can improve representation of input templates when mismatch tolerance is required [35].
Experimental Workflow for Computational Validation
The following workflow illustrates the integrated computational and experimental approach for validating primer-primer interactions:
Temperature Gradient PCR for Empirical Optimization
While computational predictions provide valuable guidance, empirical optimization remains essential for validating primer performance:
Primer Preparation: Dilute primers to working concentrations (typically 10-100 μM) and prepare master mixes containing all PCR components except templates [39].
Thermal Gradient Setup: Program a thermal cycler with a gradient of annealing temperatures, typically spanning 5-10°C below the calculated Tm of the primer pair [4].
Amplification and Analysis: Perform PCR amplification and analyze products using agarose gel electrophoresis or capillary electrophoresis to determine optimal annealing temperatures and identify non-specific products [39].
Quantitative Assessment: For qPCR applications, additionally assess amplification efficiency through standard curve analysis, with ideal primers demonstrating efficiency between 90-110% [18].
This empirical validation is particularly important for applications requiring high sensitivity and specificity, as it accounts for experimental conditions that may not be fully captured by computational models.
Research Reagent Solutions
Table 3: Essential Research Reagents for Primer Interaction Studies
| Reagent/Category | Specific Examples | Function in Primer Evaluation |
|---|---|---|
| DNA Polymerases | GoTaq Green Hot Master Mix, BioTaq DNA Polymerase | Amplification with fidelity; proofreading versions reduce errors [39] [40] |
| Modified Nucleotides | dITP, dDTP, dUTP, 5Me-dCTP | Alter hydrogen bonding properties; enable selective amplification [40] |
| Template Systems | Synthetic gBlocks, Genomic DNA, Plasmid DNA | Controlled templates for validation; known sequences for benchmarking [35] |
| Specialized Buffers | MgCl2-containing buffers, DMSO-containing buffers | Optimize hybridization stringency; reduce secondary structures [40] |
| Analysis Reagents | Ethidium Bromide, SYBR Green, Agarose | Visualize and quantify amplification products; detect primer-dimers [39] |
The integration of computational prediction tools with experimental validation provides a powerful framework for understanding and mitigating primer-primer interactions mediated by hydrogen bonding. By leveraging the capabilities of tools ranging from established standards like Primer3 to innovative machine learning approaches, researchers can significantly reduce experimental failure rates and improve the reliability of molecular assays. As these computational methods continue to evolve, incorporating more sophisticated models of hydrogen bonding dynamics and their effects on reaction kinetics, they promise to further enhance our ability to design optimal primer systems for research, diagnostic, and therapeutic applications.
Real-time quantitative PCR (RT-qPCR) and other nucleic acid amplification techniques rely heavily on fluorescent signaling for detection and quantification. The use of intercalating dyes represents a widespread, cost-effective approach for monitoring amplification in real time. However, a significant limitation of these dyes is their non-specific binding to any double-stranded DNA (dsDNA), including undesirable side products like primer-dimers, which can compromise data accuracy. The formation of these non-specific products is fundamentally governed by hydrogen bonding between complementary or partially complementary primer sequences. This technical guide explores the principles of using intercalating dyes for the detection of dimer amplification, details methodologies to identify and mitigate these artifacts, and situates this discussion within the broader research on the thermodynamics of hydrogen bonding in nucleic acid interactions.
Principles of Intercalating Dyes
Intercalating dyes, such as SYBR Green, are fluorescent molecules that emit light upon binding to the minor groove of dsDNA. In RT-qPCR, as the target DNA is amplified, the quantity of dsDNA increases with each cycle. The dye intercalates into this newly formed DNA, leading to a proportional increase in fluorescence intensity that can be measured in real-time, allowing for quantification of the initial template [41].
The key advantage of this system is its simplicity and cost-effectiveness. Unlike probe-based methods (e.g., TaqMan), which require a custom-designed, fluorescently-labeled oligonucleotide for each target, intercalating dyes can be used with any primer set, making them ideal for large-scale or rapid experimental design [41].
Hydrogen Bonding and the Genesis of Primer-Dimers
Primer-dimers are non-specific amplification artifacts formed when two primers hybridize to each other via their complementary sequences, rather than to the intended target template. The polymerase enzyme then extends the primers, creating a short, double-stranded product that can be efficiently amplified in subsequent cycles.
The initial step in dimer formation is the hydrogen bonding between the nitrogenous bases of two oligonucleotide primers. The stability of this hybridization is dictated by the number and strength of these hydrogen-bonded base pairs. This sequence-specific hydrogen bonding network is the molecular foundation upon which primer-dimer artifacts are built. Consequently, the thermodynamic stability of these unintended duplexes becomes a critical factor in their formation propensity during amplification reactions.
Experimental Detection and Analysis of Dimer Amplification
Real-Time Monitoring and Melt Curve Analysis
When using intercalating dyes, the real-time amplification plot may show a slowly rising baseline or amplification in no-template controls (NTCs), which can indicate primer-dimer formation [5]. However, the definitive diagnostic tool is the melting curve analysis (also called dissociation curve).
After the amplification cycles are complete, the temperature is gradually increased while fluorescence is continuously monitored. As the dsDNA products denature, a sharp drop in fluorescence occurs. Different DNA products melt at different temperatures based on their length, GC content, and sequence. A specific, single amplicon will produce a single, sharp peak in the melt curve. The presence of primer-dimers, which are typically shorter and have lower melting temperatures (Tm) than the specific product, will manifest as an additional, earlier peak [41].
Workflow for Melt Curve Analysis:
Impact of Dimerization on Assay Performance
The formation and amplification of primer-dimers have several detrimental effects on assay performance:
- Reduced Sensitivity: Primers sequestered into dimer complexes are unavailable for target amplification, reducing the effective primer concentration and reaction efficiency [5].
- Overestimation of Target Quantity: Fluorescence from dye binding to dimer products contributes to the total signal, leading to inaccurate quantification and lower Cq (quantification cycle) values [41].
- Increased Background Fluorescence: A steadily rising baseline in the amplification plot can obscure the early, low-amplification cycles of the specific target, making accurate Cq determination difficult [5].
Quantitative Analysis of Dimer Formation
Understanding the thermodynamic and experimental conditions that foster dimerization is key to preventing it. The following table summarizes critical findings from research on the parameters influencing primer-dimer formation.
Table 1: Quantitative Parameters for Primer-Dimer Formation
| Parameter | Threshold for Stable Dimer Formation | Experimental Context |
|---|---|---|
| Complementary Base Pairs | >15 consecutive base pairs [42] | Free-solution conjugate electrophoresis of 30-mer primers. |
| Non-consecutive Base Pairs | 20 out of 30 possible base pairs did not form stable dimers [42] | Free-solution conjugate electrophoresis. |
| Free Energy (ΔG) | More negative ΔG values correlate with higher probability of non-specific amplification [5] | RT-LAMP assay; thermodynamic modeling of primer secondary structures. |
| Temperature Dependence | Dimerization is inversely correlated with temperature [42] | Electrophoresis at temperatures from 18°C to 62°C. |
These quantitative data highlight that the spatial arrangement of complementary bases (consecutive vs. non-consecutive) is as critical as the total number of hydrogen bonds formed. Assays performed at lower temperatures, such as isothermal amplifications, are particularly susceptible [42].
The Scientist's Toolkit: Essential Reagents and Materials
Table 2: Research Reagent Solutions for Experiments with Intercalating Dyes
| Reagent/Material | Function/Description | Example Use Case |
|---|---|---|
| SYBR Green | A common intercalating dye; fluorescence increases >1000-fold upon binding dsDNA. | Real-time monitoring of dsDNA amplification in RT-qPCR [41]. |
| Bst 2.0 WarmStart Polymerase | A strand-displacing DNA polymerase for isothermal amplification (e.g., LAMP). | Used in RT-LAMP assays to study primer-dimer impact [5]. |
| Specific Primer Pairs | Oligonucleotides designed to avoid 3'-end complementarity. | The core reagent; careful design is the first line of defense against dimers. |
| Capillary Electrophoresis System | Separates DNA fragments by size to quantify dimer formation. | Used to empirically measure dimerization risk between primer-barcode pairs [42]. |
| Betaine | A zwitterionic additive that can reduce secondary structure formation. | A common component in LAMP and other amplification buffers [5]. |
Advanced Experimental Protocol: Quantifying Dimerization Risk
The following protocol, adapted from free-solution conjugate electrophoresis (FSCE) studies, provides a method to empirically quantify dimerization risk between two primers before using them in amplification assays [42].
Protocol: Mobility Shift Assay for Primer-Dimer Analysis
Primer Design and Modification:
- Design two primers suspected of forming dimers. One primer is conjugated at the 5'-end to a neutral, hydrophilic "drag-tag" (e.g., a synthetic poly-N-methoxyethylglycine) and labeled with a fluorophore (e.g., ROX). The other primer is labeled with a different fluorophore (e.g., FAM).
Annealing Reaction:
- Mix the drag-tagged and non-drag-tagged primers.
- Denature the mixture at 95°C for 5 minutes.
- Anneal by incubating at 62°C for 10 minutes, then cool to 25°C.
Free-Solution Capillary Electrophoresis (FSCE):
- Analyze the annealed mixture and single-primer controls using capillary electrophoresis without a sieving matrix.
- Use a buffer (e.g., 1x TTE) with a dynamic coating to suppress electroosmotic flow.
- Run separations at a range of temperatures (e.g., 18°C, 25°C, 40°C, 55°C, 62°C).
Data Analysis:
- The drag-tag causes a significant mobility shift, allowing clear separation of single-stranded primers from double-stranded primer-dimer complexes.
- The proportion of dimer formation is quantified by comparing the peak areas of the dimer and single-strand peaks at different temperatures.
Logical Workflow for Dimer Risk Assessment:
Intercalating dyes are a powerful tool for real-time monitoring of nucleic acid amplification, but their lack of specificity necessitates rigorous experimental design and validation. The detection of primer-dimer amplification is not merely a technical nuisance; it provides a direct window into the hydrogen bonding dynamics that govern oligonucleotide interactions. By employing quantitative tools like melt curve analysis and capillary electrophoresis, and by adhering to thermodynamic principles during primer design, researchers can distinguish specific amplification from artifactual dimerization, thereby ensuring the generation of robust and reliable quantitative data.
The formation of primer-dimers and non-specific amplification artifacts represents a significant challenge in nucleic acid diagnostics, fundamentally rooted in the predictable hydrogen bonding of the Watson-Crick framework. This technical guide explores the application of Self-Avoiding Molecular Recognition Systems (SAMRS), a class of nucleotide analogs engineered to maintain specific binding with natural DNA while avoiding mutual recognition. By strategically manipulating hydrogen bonding patterns, SAMRS technology effectively decouples primer-primer interactions from primer-template binding, offering a powerful chemical solution to the problem of cross-talk in multiplexed amplification assays. This whitepaper details the core principles, experimental implementation, and practical design considerations for deploying SAMRS to enhance assay specificity and multiplexing capability.
The specificity of nucleic acid amplification techniques, including polymerase chain reaction (PCR) and isothermal methods, is governed by the fundamental principles of molecular recognition, primarily through hydrogen bonding between complementary nucleobases. Standard DNA primers built from natural nucleotides (A, T, G, C) follow the well-established Watson-Crick rules: adenine pairs with thymine via two hydrogen bonds, and guanine pairs with cytosine via three hydrogen bonds. While this system enables specific target recognition, it also creates a vulnerability: any complementary regions between primers themselves can form stable duplexes, leading to primer-dimer (PD) formation and other non-specific amplification artifacts [3].
This cross-talk is particularly problematic in multiplexed assays, where numerous primer pairs are present simultaneously. The high local concentration of primers increases the probability of inter-primer interactions, which can consume reaction reagents, generate false-positive signals, and compete with the amplification of the desired target [5] [43]. Traditional mitigation strategies, such as hot-start enzymes and optimized thermal cycling, are often insufficient because they do not alter the inherent molecular recognition properties of the primers. The challenge, therefore, lies in the chemistry of the primers themselves. This context frames the core thesis: strategic manipulation of the hydrogen bonding patterns in nucleic acid primers can directly suppress the root cause of cross-talk, leading to more robust and highly multiplexed diagnostic assays.
SAMRS: Core Principles and Chemical Design
Self-Avoiding Molecular Recognition Systems (SAMRS) are composed of synthetic nucleotide analogs designed to form stable base pairs with their natural complements but exhibit significantly weakened binding to other SAMRS components [44] [45]. This property is achieved by redesigning the hydrogen-bonding faces of the nucleobases.
The SAMRS Alphabet and Hydrogen Bonding Logic
The first-generation SAMRS alphabet typically includes the following components, which replace their natural counterparts in oligonucleotide primers [46] [45]:
- A* (2-Aminopurine-2'-deoxyriboside): Pairs with natural T.
- T* (2'-Deoxy-2-thiothymidine): Pairs with natural A.
- G* (2'-Deoxyinosine, Hypoxanthine): Pairs with natural C.
- C* (N4-Ethyl-2'-deoxycytidine): Pairs with natural G.
The critical innovation is that the SAMRS:SAMRS pairs (A:T and G:C) are thermodynamically disfavored, forming only one weak hydrogen bond or none at all, whereas the SAMRS:natural pairs are stabilized by two hydrogen bonds, similar in strength to a natural A:T pair [46] [47]. This creates a directed binding system: SAMRS primers can bind to natural DNA templates but have a low propensity to bind to each other.
Table 1: Hydrogen Bonding Properties of SAMRS Components
| SAMRS Component | Natural Complement | Hydrogen Bonds in Pair (SAMRS:Natural) | SAMRS Complement | Hydrogen Bonds in Pair (SAMRS:SAMRS) |
|---|---|---|---|---|
| A* | T | 2 | T* | 1 |
| T* | A | 2 | A* | 1 |
| G* | C | 2 | C* | 1 |
| C* | G | 2 | G* | 1 |
The following diagram illustrates the directed binding logic of the SAMRS system, preventing primer-primer interactions while preserving primer-template binding.
Quantitative Data and Performance Analysis
Experimental data from melting temperature studies and functional PCR assays consistently demonstrate the efficacy of SAMRS in reducing cross-talk.
Thermodynamic Stability of SAMRS Complexes
Thermal denaturation studies of duplexes containing SAMRS components confirm the foundational design principle. As shown in Table 2, SAMRS nucleotides form more stable duplexes with their natural complements than with their SAMRS complements [44] [46]. For instance, a duplex with A* paired against natural T has a higher melting temperature (Tm) than a duplex with A* paired against T*. This differential stability is the thermodynamic driver of self-avoidance.
Table 2: Exemplary Melting Temperature (Tm) Data for SAMRS-Containing Duplexes
| SAMRS Component in Strand 1 | Component in Strand 2 | Observed Tm (°C) | Relative Duplex Stability |
|---|---|---|---|
| A* | T (Natural) | ~40 | High |
| A* | T* (SAMRS) | <<40 | Low |
| T* | A (Natural) | ~40 | High |
| T* | A* (SAMRS) | <<40 | Low |
| G* | C (Natural) | ~40 | High |
| G* | C* (SAMRS) | <<40 | Low |
| C* | G (Natural) | ~40 | High |
| C* | G* (SAMRS) | <<40 | Low |
Functional Performance in PCR and Isothermal Amplification
The functional benefits of SAMRS are clear in amplification experiments. In one pivotal study, a standard primer pair with perfect complementarity in their last nine 3' nucleotides failed to produce the desired amplicon and instead generated only primer-dimer. Remarkably, when both primers were synthesized with four or eight SAMRS components in their 3'-segments, the reaction efficiently produced the correct amplicon with no detectable primer-dimer [46].
This effect extends beyond PCR. For example, in Recombinase Polymerase Amplification (RPA), a low-temperature isothermal method particularly prone to off-target artifacts, the use of SAMRS-modified primers was shown to avoid most undesired side products, thereby improving the specificity and reliability of the assay [45]. Similarly, in a highly multiplexed setting targeting 14 cancer-relevant genes, chimeric primers with SAMRS components successfully amplified all targets in a single reaction, whereas standard primers failed due to cross-reactions [46].
Table 3: Impact of SAMRS on Functional Assay Performance
| Application | Assay Type | SAMRS Intervention | Key Outcome |
|---|---|---|---|
| PCR | Single-plex | 4-8 SAMRS components at 3' end of both primers | Elimination of primer-dimer; specific amplicon generated [46] |
| PCR | 10-plex | Chimeric {16 natural + 8 SAMRS + 1 natural} primers | Successful amplification of all 10 targets; control with standard primers failed [46] |
| Isothermal | RPA | SAMRS components in primers | Significant reduction of undesired side products and non-specific amplification [45] |
| SNP Detection | Allele-Specific PCR | Strategic SAMRS placement near 3' end | Improved SNP discrimination and elimination of primer-dimer artifacts [43] |
Experimental Protocol: Implementing SAMRS-Modified Primers
This section provides a detailed methodology for designing, synthesizing, and utilizing SAMRS primers in a PCR assay, based on established protocols [46] [43].
Primer Design and Synthesis
- Sequence Selection: Begin with a standard primer sequence designed for your target using conventional software (e.g., Primer-BLAST). The initial sequence should be 18-24 nucleotides in length with a Tm of 54-65°C and GC content of 40-60% [18].
- Strategic SAMRS Incorporation:
- Positioning: Replace standard nucleotides with their SAMRS counterparts (A→A, T→T, G→G, C→C) strategically in the 3'-segment of the primer. The most critical positions are those involved in potential primer-primer complementarity.
- Architecture: A common and effective design is the chimeric "{X + Y* + 1}" architecture, where:
- Completeness: For maximum self-avoidance, both the forward and reverse primers in a pair should be modified with SAMRS.
- Synthesis and Purification: SAMRS-containing oligonucleotides are synthesized using standard phosphoramidite chemistry. The SAMRS phosphoramidites (commercially available from suppliers like Glen Research or ChemGenes) couple with standard protocols. Post-synthesis, purify the primers by ion-exchange HPLC to a purity of >85-90% [43].
PCR Reaction Setup and Thermal Cycling
The following protocol is adapted for use with SAMRS primers and Taq DNA polymerase, which has been shown to efficiently incorporate and read through SAMRS components [46].
Table 4: Research Reagent Solutions for SAMRS-PCR
| Reagent | Function | Notes/Specification |
|---|---|---|
| SAMRS-Modified Primers | Sequence-specific amplification | Chimeric design (e.g., {16+8*+1}); HPLC purified [43] |
| Taq DNA Polymerase | DNA synthesis | Preferable over other thermostable polymerases for better efficiency with SAMRS [46] |
| dNTP Mix | Nucleotide substrates | Standard dNTP mixture (e.g., 1.4 mM each) |
| Reaction Buffer | Optimal enzyme activity | 1x concentration, may require Mg++ supplementation to 5.0 mM [43] |
| RNase-Free Water | Reaction solvent | Nuclease-free to prevent oligonucleotide degradation |
Procedure:
- Prepare Master Mix (for a 25 µL reaction):
- 1X PCR Buffer (e.g., 10 mM Tris-HCl, 50 mM KCl, pH 8.3)
- 1.5-5.0 mM MgCl₂ (optimization may be required)
- 200 µM of each dNTP
- 0.2-0.5 µM of each SAMRS-modified forward and reverse primer
- 0.5-1.25 U of Taq DNA Polymerase
- Nuclease-free water to volume
- Add Template DNA: Add 1-100 ng of genomic DNA or equivalent template.
- Thermal Cycling:
- Initial Denaturation: 95°C for 3 min.
- Amplification (35-40 cycles):
- Denature: 95°C for 30 sec.
- Anneal: The annealing temperature (Ta) is critical. Start with a Ta 2-5°C below the calculated Tm of the SAMRS:natural duplex. Note: The Tm of a SAMRS primer bound to its natural DNA target will be lower than that of a standard primer of the same sequence. Empirical optimization is recommended.
- Extend: 72°C for 1 min per kb.
- Final Extension: 72°C for 5 min.
- Analysis: Analyze PCR products by agarose gel electrophoresis. The expected outcome is a clear band of the desired amplicon with a significant reduction or absence of low-molecular-weight primer-dimer artifacts compared to a control reaction with standard primers.
The following workflow summarizes the key steps in the SAMRS primer design and experimental process.
Discussion and Design Considerations
Advantages and Limitations
The primary advantage of SAMRS is its ability to chemically enforce specificity, moving beyond software-based design to directly alter the molecular interactions between primers. This is especially valuable in complex applications like high-level multiplex PCR (20-plex and beyond) [45], SNP detection where 3'-end specificity is paramount [43], and low-temperature isothermal amplifications like RPA [45].
A key consideration is the reduced binding strength of SAMRS:natural pairs compared to natural:natural pairs. Since each SAMRS:natural pair is stabilized by only two hydrogen bonds, the overall Tm of a SAMRS primer will be lower than that of its all-natural counterpart. This necessitates careful design and may require lowering the PCR annealing temperature. Furthermore, the synthesis cost of SAMRS oligonucleotides is higher than for standard primers, and not all DNA polymerases process SAMRS components with equal efficiency, requiring empirical validation [46].
Comparison with Alternative Technologies
SAMRS represents a distinct approach compared to other dimer-suppression technologies. For example, rhPCR (RNase H-dependent PCR) uses primers with a single RNA base and a 3' blocker. These primers are only activated after cleavage by RNase H2, which occurs efficiently only when the primer is perfectly hybridized to its target. This also effectively reduces primer-dimer formation but requires an additional enzyme and a different primer design paradigm [48]. While both are effective, SAMRS acts at the level of fundamental molecular recognition, whereas rhPCR acts through enzymatic control of primer activation.
SAMRS technology provides a powerful and direct solution to the problem of cross-talk in nucleic acid amplification by addressing its root cause: the promiscuous hydrogen bonding of natural nucleotides. By designing primers that follow a self-avoiding recognition code, researchers can significantly suppress primer-dimer formation and non-specific amplification. This enables more robust, sensitive, and highly multiplexed assays for advanced diagnostics, pathogen detection, and genotyping. As the field of synthetic biology continues to provide new tools, the integration of advanced molecular systems like SAMRS will be crucial for pushing the boundaries of what is possible in molecular diagnostics.
Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) has emerged as a powerful molecular tool for the rapid detection of RNA viruses, particularly in resource-limited settings. This case study explores the critical process of primer redesign for Dengue Virus (DENV) and Yellow Fever Virus (YFV) detection, framing the discussion within the broader context of hydrogen bonding interactions and their role in primer dimer formation. The performance of RT-LAMP assays is profoundly influenced by primer design, which directly affects specificity, sensitivity, and amplification efficiency. Hydrogen bonding stability between primer and template, as well as between primers themselves, represents a fundamental molecular interaction that can determine assay success or failure. This technical guide provides a comprehensive framework for researchers seeking to optimize RT-LAMP assays for these clinically significant flaviviruses, with particular emphasis on the molecular interactions that underpin primer functionality.
Technical Foundations of RT-LAMP
Core Principles and Advantages
RT-LAMP combines reverse transcription of RNA templates with DNA amplification under isothermal conditions (typically 60-65°C) using a strand-displacing DNA polymerase, most commonly from Bacillus stearothermophilus (Bst) [49] [50]. Unlike conventional PCR-based methods that require thermal cycling, RT-LAMP maintains a constant temperature, eliminating the need for sophisticated instrumentation [51]. The technique employs four to six primers targeting six to eight distinct regions of the target sequence, which confers exceptional specificity and enables rapid amplification within 15-60 minutes [52] [53].
Key advantages of RT-LAMP include:
- Rapid results (typically 30-60 minutes compared to 2+ hours for RT-qPCR)
- Isothermal conditions (simplifying instrumentation requirements)
- High tolerance to inhibitors (enabling simplified sample preparation)
- Multiple detection modalities (colorimetric, fluorescent, turbidimetric, lateral flow)
- Excellent sensitivity and specificity comparable to RT-qPCR [51] [54]
Primer Architecture in RT-LAMP
A complete RT-LAMP primer set typically consists of the following components:
- F3 and B3 (Forward and Backward Outer Primers): Short primers (18-22 nt) that initiate strand displacement
- FIP and BIP (Forward and Backward Inner Primers): Longer primers (40-45 nt) containing complementary sequences that form loop structures
- LF and LB (Loop Forward and Loop Backward Primers): Optional primers that accelerate reaction kinetics by binding to loop regions [55] [52]
The strategic arrangement of these primers enables the formation of characteristic stem-loop DNA structures that drive continuous amplification without denaturation steps. This complex primer architecture, while enabling rapid amplification, also increases the potential for off-target interactions mediated by hydrogen bonding, particularly when primers are poorly designed.
Primer Design Strategies for Flaviviruses
Target Selection and Conservation Analysis
Effective primer redesign begins with identification of appropriate genomic targets. For flaviviruses like DENV and YFV, conserved regions across serotypes or genotypes are essential for broad detection capabilities.
Table 1: Target Genes for DENV and YFV RT-LAMP Primer Design
| Virus | Target Genes | Conservation Considerations | Reported Sensitivity |
|---|---|---|---|
| Dengue | 3' UTR, 5' UTR, NS2A, NS4A, NS4B | Pan-serotype assays require regions conserved across DENV 1-4 | 86.3-98.9% [53] [56] |
| Yellow Fever | NS5, E, NS1 | Targets must distinguish between wild-type and vaccine strains (17D) | 100% [50] [57] |
For DENV, the 5' and 3' untranslated regions (UTRs) demonstrate high conservation across serotypes, making them ideal targets for pan-serotype detection [51]. The non-structural proteins NS2A, NS4A, and NS4B have also been successfully employed in serotype-specific assays [56]. For YFV, the NS5 and envelope (E) genes contain highly conserved regions suitable for primer design [50], while the NS1 gene has also been successfully targeted [57].
Sequence Alignment and Conservation Mapping
Multiple sequence alignment using tools like Clustal Omega or MUSCLE is essential to identify conserved regions suitable for primer binding [50] [55]. The alignment should encompass diverse viral strains, including:
- All four DENV serotypes for pan-serotype assays
- Contemporary and historical YFV strains
- Geographic variants from different outbreak regions
- Vaccine strains (e.g., YFV 17D) if discrimination is required
SimPlot analysis can graphically represent sequence conservation, revealing regions of high homology ideal for primer targeting [55]. For DENV, this approach has identified highly conserved sequences in the 5' and 3' UTRs with minimal secondary structure, facilitating primer binding [51].
Primer Design Parameters and Optimization
Table 2: Optimal Parameters for RT-LAMP Primer Design
| Parameter | Optimal Range | Functional Significance |
|---|---|---|
| Length (F3/B3) | 18-22 bp | Determines binding specificity and hydrogen bonding stability |
| Length (FIP/BIP) | 40-45 bp | Encompasses F1c+F2 or B1c+B2 regions with TTTT linker |
| GC Content | 40-60% | Balances hybridization stability and prevents secondary structures |
| Tm F1c/B1c | 63-65°C | Critical for loop formation and strand displacement |
| Tm F2/B2/F3/B3 | 58-61°C | Ensures simultaneous binding at reaction temperature |
| ΔG (3' end) | ≤ -4 kcal/mol | Prevents nonspecific initiation and primer dimerization |
| Distance F2-B2 | 120-160 bp | Optimal amplicon size for efficient amplification |
| Distance F2-F1 | 40-60 bp | Proper spacing for loop formation |
Advanced design considerations include:
- Incorporation of degenerate bases to account for sequence variability while maintaining hydrogen bonding potential [50]
- TTTT linker sequences between F1c and F2 (and B1c and B2) components of inner primers to improve hybridization sensitivity [50]
- Explicit setting of ΔG values for 3' ends to prevent nonspecific amplification [55]
- Avoidance of continuous GC-rich stretches that promote stable mispriming through excessive hydrogen bonding
Experimental Validation Protocols
Specificity Testing
Comprehensive specificity validation must include:
- Cross-reactivity assessment with related flaviviruses (Zika, West Nile, Japanese encephalitis) [50] [56]
- Serotype differentiation for DENV assays [56]
- Wild-type vs vaccine strain discrimination for YFV [57]
- Human genomic DNA and common respiratory pathogens to exclude false positives
Testing should be performed in at least ten independent replicates for each control to establish robust specificity metrics [50]. For DENV assays, this process has demonstrated 100% specificity with no cross-reactivity between serotypes when properly designed [56].
Sensitivity and Limit of Detection
Sensitivity validation requires:
- RNA transcript dilution series from known copy numbers
- Clinical isolate serial dilutions in relevant matrices
- Comparison with reference methods (RT-qPCR, plaque assay)
- Probit analysis for statistical determination of limit of detection [50]
Properly designed DENV RT-LAMP assays have demonstrated detection limits of 10-20 RNA copies per reaction, representing a 10-fold improvement over conventional RT-PCR in some cases [51] [56]. For YFV, assays have achieved detection limits as low as 12 PFU/mL [50].
Clinical Performance Evaluation
Clinical validation should include:
- Blinded testing of characterized clinical specimens
- Calculation of sensitivity, specificity, PPV, and NPV with confidence intervals
- Concordance analysis with gold standard methods (kappa statistic)
- Inclusivity testing across diverse geographic isolates
In clinical settings, redesigned DENV RT-LAMP assays have shown sensitivity of 86.3-95% and specificity of 93-99% compared to RT-qPCR [51] [53]. For YFV, redesigned assays demonstrated 100% sensitivity and specificity compared to RT-qPCR in testing with non-human primate samples [50].
Addressing Hydrogen Bonding in Primer Dimers
Molecular Mechanisms of Primer Dimerization
Primer dimers form through interspecific hydrogen bonding between complementary sequences in primers, particularly at 3' ends. In RT-LAMP, the large number of primers (4-6) significantly increases the potential for these interactions. The stable hydrogen bonding networks that form between mispaired primers can initiate non-template-dependent amplification, consuming reagents and generating false-positive signals.
Key factors contributing to primer dimer formation include:
- 3' complementarity between primers (even 2-3 bases can initiate extension)
- GC-rich regions with excessive hydrogen bonding potential
- Secondary structures in primers that expose complementary sequences
- Suboptimal reaction temperatures that allow transient hybridization
Computational Prediction and Mitigation
Advanced primer design strategies to minimize hydrogen bonding issues include:
- Explicit calculation of ΔG dimer values (> -2 kcal/mol recommended) [55]
- Secondary structure prediction at operational temperatures [55]
- Incorporation of molecular switches (temperature-sensitive primers) [54]
- Reduced primer sets (5 primers instead of 6) to decrease interaction complexity [52]
The use of five-primer systems (omitting one loop primer) has demonstrated significant reduction in false-positive rates while maintaining high sensitivity [52]. This approach decreases the combinatorial complexity of potential primer interactions while only marginally reducing amplification kinetics.
Experimental Confirmation
Wet-lab validation of primer dimer formation includes:
- Extended incubation (up to 120 minutes) to detect late false amplification [52]
- No-template controls with multiple replicates
- Gel electrophoresis to distinguish specific amplicons from primer dimers
- Melting curve analysis for amplicon characterization
Properly designed primers should show no amplification in NTCs even after extended incubation periods, demonstrating minimal problematic hydrogen bonding between primers [52].
Primer Redesign and Validation Workflow
Research Reagent Solutions
Table 3: Essential Reagents for RT-LAMP Assay Development
| Reagent Category | Specific Examples | Function and Application Notes |
|---|---|---|
| Polymerase Enzymes | Bst 2.0, Bst 3.0 DNA Polymerase | Strand-displacing activity with reverse transcriptase capability in Bst 3.0 |
| Master Mixes | WarmStart Colorimetric LAMP 2X Master Mix | Includes pH-sensitive dye for visual detection, contains required buffers and dNTPs |
| Enhancement Additives | Guanidine Hydrochloride (GuHCl) | Improves amplification efficiency and reduces detection time by 22% [52] |
| Primer Design Tools | PrimerExplorer V5, NEB LAMP Primer Design Tool | Specialized algorithms for LAMP primer design with parameter optimization |
| Detection Systems | Phenol Red, SYBR Green, Calcein, Lateral Flow Strips | Colorimetric, fluorescent, or immunochromatographic result interpretation |
| Sample Preparation | Quick-RNA Viral Kit, Blood Lysis Buffer (Triton X-100) | RNA extraction or simplified processing for field applications [51] |
Case Studies in Primer Redesign
Dengue Virus Pan-Serotype Detection
A significant challenge in DENV diagnosis is simultaneous detection of all four serotypes with uniform sensitivity. Traditional approaches targeting structural genes showed variable performance across serotypes. Redesigned primers incorporating the following improvements demonstrated enhanced performance:
- Combined 5' and 3' UTR targeting in a single reaction (COMB-RT-LAMP) increased sensitivity from <40% to >95% [51]
- Strategic placement of degenerate bases to accommodate serotype variability while maintaining hydrogen bonding stability
- Optimized inner primer length to balance binding energy and specificity
The redesigned assay detected all four serotypes with equal efficiency and demonstrated 95% sensitivity in clinical validation with febrile patients from Colombia [51].
Yellow Fever Virus Surveillance
Primer redesign for YFV focused on distinguishing circulating wild-type strains while maintaining detection of diverse genotypes. The redesign process included:
- Comprehensive analysis of 50 complete YFV genomes from Brazilian isolates [50]
- NS5 and E gene targeting with degenerate primers to ensure broad detection
- Exclusion of vaccine strain sequences from conservation analysis when discrimination was desired
The validated assay demonstrated 100% sensitivity and specificity with a detection limit of 12 PFU/mL, suitable for surveillance of non-human primate samples in Brazil [50].
Hydrogen Bonding Interactions in Primer Function
The redesign of RT-LAMP primers for dengue and yellow fever virus detection represents a sophisticated interplay between molecular biology, bioinformatics, and biochemical principles. The strategic management of hydrogen bonding interactions through careful primer design is fundamental to achieving optimal assay performance. By applying the methodologies and validation frameworks outlined in this technical guide, researchers can develop robust, reliable detection systems suitable for both clinical diagnosis and field surveillance. The continued refinement of primer design strategies, with particular attention to the molecular interactions that govern specificity, will further enhance the utility of RT-LAMP technology in global infectious disease monitoring.
Solving the Dimer Dilemma: A Troubleshooting Guide for Robust PCR
In the polymerase chain reaction (PCR), the annealing step is a critical determinant of success, governed fundamentally by the thermodynamics of hydrogen bonding between primers and their target DNA sequences. The annealing temperature must be precisely optimized to facilitate specific primer-template hybridization while minimizing non-specific interactions that lead to inefficient amplification and artifacts such as primer-dimers.
Primer-dimers, a common PCR artifact, are short, double-stranded DNA fragments formed by the interaction between two primers. Their formation is driven by hydrogen bonding between complementary bases, particularly stable G-C pairs with three hydrogen bonds, which can outcompete desired primer-template binding under suboptimal conditions. This guide details the optimization of thermal cycler conditions, focusing on annealing temperature selection and advanced protocols like touchdown PCR to suppress such artifacts and achieve high-specificity amplification.
Primer Design and the Biochemistry of Hydrogen Bonding
Successful PCR amplification begins with well-designed primers. The physical properties of primers, dictated by their sequence, directly influence their interaction with the template and with each other via hydrogen bonding.
Table 1: Optimal Primer Design Parameters [58] [25] [31]
| Parameter | Optimal Range or Value | Rationale and Impact on Hydrogen Bonding |
|---|---|---|
| Length | 18–30 nucleotides | Determines the total number of potential hydrogen bonds for stability. |
| Melting Temperature (Tm) | 55–72°C; within 5°C for primer pair | Direct reflection of cumulative hydrogen bond strength in primer-template duplex. |
| GC Content | 40–60% | Higher GC content increases Tm due to more G-C pairs (3 H-bonds each). |
| 3' End Stability | 2–3 G or C bases | Strong terminal hydrogen bonding promotes specific initiation of extension. |
| Self-Complementarity | ≤3 contiguous bases | Minimizes intra-primer hydrogen bonding that causes hairpins. |
| Inter-Primer Complementarity | ≤3 contiguous bases | Minimizes inter-primer hydrogen bonding that causes primer-dimers. |
The stability of the primer-template duplex is quantified by its melting temperature (Tm). A common formula for estimating Tm is: Tm = 4 °C x (G + C) + 2 °C x (A + T) [25] This calculation underscores that G-C base pairs, with three hydrogen bonds, contribute twice as much to duplex stability as A-T pairs, which have only two. Mismatches in the primer-template duplex reduce the number of hydrogen bonds, destabilizing the interaction. This principle is leveraged in touchdown PCR, where initial high-temperature annealing favors the formation of perfect, stable matches.
Quantitative Optimization of Thermal Cycler Conditions
Optimizing thermal cycler parameters is essential for balancing yield, specificity, and fidelity. The following tables summarize key reagent concentrations and standard cycling conditions.
Table 2: Key PCR Reagent Concentrations for a 50 μL Reaction [58] [59]
| Reagent | Typical Stock Concentration | Final Concentration in Reaction |
|---|---|---|
| MgCl₂ | 25 mM | 1.5 - 2.0 mM (optimize 1.0-4.0 mM) |
| dNTPs | 10 mM each | 200 μM each |
| Forward/Reverse Primers | 20 μM each | 0.1 - 1.0 μM each |
| DNA Template | Variable | ~10⁴ - 10⁶ copies (e.g., 10-100 ng human gDNA) |
| Taq DNA Polymerase | 5 U/μL | 1.25 - 2.5 U |
Table 3: Standard Three-Step PCR Cycling Conditions [58] [59]
| Step | Temperature | Time | Notes |
|---|---|---|---|
| Initial Denaturation | 94–98°C | 1–5 minutes | Duration depends on polymerase and template complexity. |
| Denaturation | 94–98°C | 10–60 seconds | |
| Annealing | Tm -5°C to Tm | 30–60 seconds | Temperature is key for specificity; see Section 3.1. |
| Extension | 68–72°C | 1 min/kb | Time depends on polymerase processivity and amplicon length. |
| Final Extension | 68–72°C | 5–10 minutes | |
| Cycle Number | 25–35 | Too many cycles can increase non-specific products. |
Empirical Determination of Annealing Temperature
The theoretical annealing temperature (Ta) can be calculated, but empirical determination is superior. A gradient PCR is the definitive method for identifying the optimal Ta [25]:
- Setup: Prepare identical PCR mixtures and run them in a thermal cycler with a defined annealing temperature gradient across the block.
- Analysis: After amplification, analyze the products by agarose gel electrophoresis.
- Selection: The annealing temperature that produces the brightest, single band of the expected size with the least background smearing or non-specific bands is the optimal Ta. This temperature is typically 5–10°C below the Tm of the primers [25].
Touchdown PCR: A Protocol for Enhanced Specificity
Touchdown PCR is a powerful modification designed to increase amplification specificity and sensitivity by progressively lowering the annealing temperature during the early cycles of the reaction [60] [61]. This method favors the accumulation of the desired specific product early on, which then outcompetes non-specific products in later cycles.
Mechanism and Workflow
The protocol begins with an annealing temperature 5–10°C above the calculated Tm of the primers. Under these stringent conditions, only primers with perfect complementarity to the template can form stable duplexes (with a maximum number of hydrogen bonds) and initiate extension. Over subsequent cycles, the annealing temperature is incrementally decreased (e.g., by 0.5–1°C per cycle) until it reaches a final, permissive temperature below the Tm.
The specific product, amplified in the initial cycles, has a numerical advantage and is preferentially amplified in later cycles, even at lower, less stringent temperatures. This process effectively reduces mispriming and primer-dimer formation.
Diagram: Touchdown PCR uses progressively lower annealing temperatures to favor specific products that form stable hydrogen bonds early.
Detailed Touchdown PCR Protocol
The following is a generalized touchdown PCR protocol based on a primer pair with a calculated Tm of 57°C [60].
Table 4: Example Touchdown PCR Protocol [60]
| Step | Temperature (°C) | Time | Number of Cycles | Stage Description |
|---|---|---|---|---|
| 1. Initial Denaturation | 95 | 3:00 | 1 | - |
| 2. Denaturation | 95 | 0:30 | 10 | Touchdown Phase: |
| 3. Annealing | 67 (Tm+10) | 0:45 | 1 | Start 10°C above Tm. |
| 4. Extension | 72 | 0:45 | 1 | - |
| 5. Denaturation | 95 | 0:30 | 15-20 | Amplification Phase: |
| 6. Annealing | 57 (Tm) | 0:45 | 1 | Use final touchdown temperature. |
| 7. Extension | 72 | 0:45 | 1 | - |
| 8. Final Extension | 72 | 5:00 | 1 | - |
Key Tips for Success [60]:
- Keep Reactions Cool: Assemble reactions on ice to prevent non-specific priming before cycling begins.
- Use Hot-Start Polymerase: This prevents primer-dimer formation and non-specific extension during reaction setup.
- Limit Total Cycles: Keep the total number of amplification cycles (touchdown + standard) below 35 to minimize non-specific product appearance.
- Combine with Additives: For difficult templates (e.g., GC-rich), combine touchdown PCR with additives like DMSO or betaine.
Special Considerations for Challenging Templates
GC-Rich Templates
GC-rich sequences (>65% GC content) are challenging due to their propensity to form stable secondary structures via strong intramolecular hydrogen bonding (e.g., hairpins) and their high duplex stability [62].
- Higher Denaturation Temperature: Use 98°C for complete strand separation [59] [62].
- Polymerase Choice: Use polymerases specifically formulated for GC-rich templates, often supplied with specialized "GC Buffers" or enhancers [62].
- Additives: Incorporate DMSO (1-10%), formamide (1.25-10%), or betaine. These additives weaken hydrogen bonding, helping to denature secondary structures and lower the effective Tm [58] [59] [62].
- Touchdown PCR: This protocol is highly recommended for GC-rich targets [61].
Preventing Primer-Dimer Formation
Primer-dimers form when primers anneal to each other via complementary sequences, particularly at their 3' ends, allowing DNA polymerase to extend the duplex. Experimental studies show that stable primer-dimer formation requires more than 15 consecutive complementary base pairs between primers [42]. Non-consecutive base pairs, even if 20 out of 30 are complementary, do not typically form stable dimers, highlighting the critical importance of contiguous hydrogen bonding for stability [42].
Diagram: Primer-dimer formation requires sufficient consecutive complementary bases for stable hydrogen bonding, enabling polymerase extension.
Prevention strategies focus on primer design and reaction conditions [58] [25] [31]:
- Primer Design: Ensure primers lack self-complementarity and 3'-end complementarity. Use primer analysis software to check for dimer potential.
- Optimal Primer Concentration: High primer concentrations increase the chance of primer-primer interactions.
- Hot-Start PCR: Employ hot-start polymerases to inhibit activity at low temperatures during setup.
- Optimize Mg²⁺ Concentration: Excess Mg²⁺ can stabilize non-specific duplexes and promote primer-dimer formation [59].
Table 5: Research Reagent Solutions for PCR Optimization
| Item | Function/Benefit | Example Use Case |
|---|---|---|
| Hot-Start DNA Polymerase | Prevents non-specific amplification and primer-dimer formation during reaction setup by requiring heat activation. | Essential for high-specificity assays and low-template PCR [58] [60]. |
| High-Fidelity Polymerase | Possesses 3'→5' exonuclease (proofreading) activity for high-fidelity amplification, crucial for cloning and sequencing. | Q5 High-Fidelity DNA Polymerase, Pfu Polymerase [58] [62]. |
| GC Enhancer / Additives | Chemical additives that disrupt secondary structures and modulate hydrogen bonding stability. | DMSO, Betaine, Formamide for GC-rich templates [58] [62]. |
| MgCl₂ Solution | Separate Mg²⁺ stock allows for fine-tuning of this critical cofactor concentration to optimize specificity and yield. | Titrating from 1.0 mM to 4.0 mM to find the optimal concentration [59] [62]. |
| Gradient Thermal Cycler | Allows a range of temperatures to be tested across a single block, enabling empirical determination of optimal Ta. | Found in most modern thermal cyclers; essential for protocol optimization [25]. |
| Primer Design Software | Algorithms (e.g., Primer3, Primer-BLAST) automate design and check for secondary structures and dimer potential. | NCBI Primer-BLAST for specific primer design and in-silico specificity checking [25] [31]. |
The precision of in vitro reactions, particularly the Polymerase Chain Reaction (PCR), is a cornerstone of modern molecular biology, diagnostics, and drug development. The success of these reactions hinges on a delicate balance of chemical components that stabilize DNA polymerases, facilitate primer-template binding, and ensure the high-fidelity synthesis of specific amplification products. Among these components, the divalent cation Mg²⁺ and chemical additives like dimethyl sulfoxide (DMSO) and betaine are particularly crucial. Their concentrations can dramatically influence reaction efficiency, especially when challenging templates such as those with high GC-content or complex secondary structures are involved.
This guide frames the discussion of these reagents within a critical research context: understanding and mitigating the role of hydrogen bonding in the formation of primer dimers. Primer dimers are a common artifact in PCR where primers hybridize to each other instead of the template DNA, largely driven by complementary sequences that form stable hydrogen-bonded networks. This non-productive pathway consumes reagents, reduces the yield of the desired product, and compromises assay sensitivity. The strategic use of Mg²⁺, DMSO, and betaine allows researchers to manipulate the hydrogen bonding landscape and reaction kinetics, thereby suppressing these aberrant pathways and enhancing the specificity and yield of target amplification for robust and reliable results [63] [64] [65].
The Fundamental Roles of Key Reaction Components
Each critical component in a PCR mixture performs a unique and essential function. Understanding their individual mechanisms of action is a prerequisite for effectively fine-tuning their interactions.
Mg²⁺: The Essential Cofactor: Magnesium ions (Mg²⁺) are an absolute requirement for DNA polymerase activity. They serve as a cofactor that facilitates the nucleophilic attack of the 3' hydroxyl group of the primer on the alpha-phosphate of the incoming dNTP, enabling the formation of the phosphodiester bond. The concentration of free Mg²⁺ is critical; it must be sufficient to saturate the dNTPs (which also chelate Mg²⁺) and form a functional complex with the polymerase. However, excess Mg²⁺ can reduce fidelity by stabilizing non-specific primer-template interactions and can even promote non-specific amplification [64] [65].
DMSO: A Secondary Structure Disruptor: Dimethyl sulfoxide (DMSO) is a polar aprotic solvent that interferes with the hydrogen bonding network of DNA. By binding in the major and minor grooves of the DNA double helix, DMSO destabilizes the DNA duplex, effectively lowering its melting temperature (T(_m)). This is particularly beneficial for denaturing stable secondary structures and hairpins in GC-rich templates that would otherwise impede polymerase progression. A significant caveat is that DMSO can also inhibit Taq polymerase activity, necessitating empirical optimization of its concentration [63] [65].
Betaine: A DNA Homogenizer: Betaine (N,N,N-trimethylglycine) is a zwitterionic osmolyte that enhances the amplification of GC-rich templates through a unique mechanism. It accumulates in the minor groove of DNA and homogenizes the thermodynamic stability of GC-rich and AT-rich regions. This equalization prevents the localized, premature denaturation of AT-rich tracts and promotes the uniform denaturation of the entire template, thereby improving the yield and specificity of long-range and difficult PCRs [63] [64].
The Hydrogen Bonding Link to Primer Dimer Formation
Primer dimer formation is a direct consequence of unintended hydrogen bonding between primer molecules. The following diagram illustrates the competitive pathways of specific amplification versus primer dimerization, highlighting the points where Mg²⁺, DMSO, and betaine exert their influence.
Diagram 1: Competitive pathways of specific amplification and primer dimer formation, showing intervention points for Mg²⁺ and additives.
As shown in Diagram 1, the reaction can proceed down one of two competing paths. The desired path involves primers forming a stable, fully complementary hydrogen-bonded network with the target template DNA, leading to specific amplification. The undesirable path occurs when the 3' ends of primers transiently form partial, low-stringency hydrogen bonds with each other. If stabilized, particularly by excess Mg²⁺, these primer-primer complexes can be extended by the polymerase, forming primer dimers that deplete reaction resources [64] [65]. Additives like DMSO and betaine can help steer this equilibrium toward the desired path by altering the hydrogen bonding efficiency and the thermal stability of these initial, non-specific interactions.
Quantitative Guide to Reagent Optimization
The table below provides a consolidated overview of the three key reagents discussed, their primary mechanisms, and their optimal concentration ranges for experimental optimization.
Table 1: Quantitative Summary of Key PCR Reagents for Optimization
| Reagent | Primary Mechanism of Action | Typical Working Concentration | Impact on Primer Dimers |
|---|---|---|---|
| Mg²⁺ | Essential cofactor for DNA polymerase; stabilizes DNA duplex [64] [65] | 1.0 - 4.0 mM (titrate in 0.5 mM steps) [64] [65] | High concentrations promote dimer formation by stabilizing transient primer-primer interactions [65]. |
| DMSO | Disrupts DNA secondary structure by interfering with hydrogen bonding; lowers template T(_m) [63] [65] | 2 - 10% (v/v) [64] [65] | Can reduce dimers by disrupting H-bonds in primer-primer complexes, but can inhibit polymerase at high concentrations [65]. |
| Betaine | Homogenizes DNA melting temperatures; equalizes stability of GC and AT base pairs [63] [64] | 1.0 - 1.7 M [65] | Can reduce dimers by reducing base-pair composition dependence of DNA melting, preventing aberrant initiation [63]. |
Experimental Protocol for Systematic Optimization
Fine-tuning a reaction requires a systematic approach. The following workflow provides a step-by-step methodology for empirically determining the optimal concentrations of Mg²⁺, DMSO, and betaine for a specific assay.
Diagram 2: A sequential workflow for the empirical optimization of Mg²⁺ and additive concentrations.
Step-by-Step Procedure:
Mg²⁺ Titration: Prepare a master reaction mix containing all standard components (buffer, dNTPs, primers, template, polymerase). Aliquot the mix and supplement with MgCl₂ or MgSO₄ to create a series of reactions with final Mg²⁺ concentrations ranging from 1.0 mM to 4.0 mM in increments of 0.5 mM [64] [65]. Run the reactions using a standard thermal cycling protocol.
Initial Analysis: Analyze the amplification products using agarose gel electrophoresis. Identify the Mg²⁺ concentration that yields the strongest target band with the least background smearing or spurious bands, which may include primer dimers (visible as a low molecular weight smear near the well front) [64].
Additive Screening: If specificity or yield remains unsatisfactory, repeat the process using the optimal Mg²⁺ concentration as the new baseline. Test the addition of DMSO and betaine, both individually and in combination.
Final Validation: Once a promising condition is identified (e.g., 2.5 mM Mg²⁺ and 3% DMSO), perform replicate reactions to confirm robustness. The final validation should assess not just amplicon yield and purity, but also fidelity, especially for applications like cloning where sequence accuracy is paramount [64].
The Scientist's Toolkit: Essential Research Reagents
A successful optimization experiment requires high-quality starting materials and reagents. The following table lists key solutions and materials essential for this work.
Table 2: Essential Reagents and Materials for Reaction Optimization
| Reagent / Material | Function / Purpose | Key Considerations |
|---|---|---|
| High-Fidelity DNA Polymerase | Enzymatic synthesis of DNA with proofreading (3'→5' exonuclease) activity for superior accuracy [64]. | Lower error rate (e.g., 10⁻⁶ vs 10⁻⁴ for Taq) is critical for cloning and sequencing [64]. |
| Molecular Biology Grade Water | Solvent for all reaction components; must be nuclease-free to prevent degradation of primers and template. | A common source of failed reactions if contaminated. |
| dNTP Mix | Building blocks (dATP, dCTP, dGTP, dTTP) for DNA synthesis. | Concentration must be balanced; dNTPs chelate Mg²⁺, affecting free Mg²⁺ availability [65]. |
| MgCl₂ or MgSO₄ Stock Solution | Source of essential Mg²⁺ cofactor. | Fully thaw and vortex stock before use to avoid concentration gradients from freeze-thaw cycles [65]. |
| Ultra-Pure Template DNA | The target DNA to be amplified. | Should be free of inhibitors like phenol, EDTA, or heparin which chelate Mg²⁺ and inhibit polymerase [64]. |
| Optimized Primer Pairs | Short, single-stranded DNA sequences that define the start and end of the amplification target. | Designed with optimal length (18-24 bp), Tm (55-65°C), and GC content (40-60%) to minimize dimerization [64]. |
Advanced Applications and Future Directions
The principles of chemical enhancement extend beyond basic PCR and are vital for advanced techniques. Long-range PCR, which involves amplifying fragments over several kilobases, heavily relies on additive cocktails to overcome the challenges associated with replicating long, complex templates [63]. Similarly, the amplification of GC-rich templates (>65% GC) is notoriously difficult due to the formation of stable secondary structures; this is a classic scenario where the combination of a lowered Mg²⁺ concentration, 2-10% DMSO, and 1-1.7 M betaine can make the difference between amplification failure and success [63] [64] [65].
Future research will continue to refine our understanding of how reagents like betaine and DMSO alter the fundamental thermodynamics of nucleic acid hybridization. Furthermore, the development of proprietary enhancer cocktails by commercial entities demonstrates the move towards empirically optimized, pre-mixed solutions designed to tackle a wide array of challenging amplification scenarios, making optimized reaction chemistry more accessible to researchers [63]. The insights gained from fine-tuning these in vitro reactions also contribute to a deeper understanding of in vivo biomolecular interactions, including the principles of hydrogen bonding and its role in genetic regulation and disease mechanisms.
In molecular biology research and drug development, the accuracy of polymerase chain reaction (PCR) is paramount. High-fidelity DNA polymerases are specialized enzymes designed to amplify target DNA sequences with exceptional accuracy, minimizing the introduction of errors during amplification. These enzymes achieve superior fidelity primarily through proofreading activity, a 3'→5' exonuclease function that corrects misincorporated nucleotides during DNA synthesis [66]. For applications such as cloning, sequencing, and gene expression where precise DNA sequence integrity is critical, high-fidelity polymerases are indispensable tools.
The fundamental importance of polymerase selection extends into basic research on molecular interactions, particularly hydrogen bonding in primer dimers. The formation of primer dimers—where primers hybridize to each other rather than the target template—occurs through complementary base pairing stabilized by hydrogen bonds [3]. Understanding how different polymerase enzymes influence or are affected by these non-target hydrogen bonding interactions is crucial for optimizing PCR specificity and efficiency, especially when working with challenging templates such as GC-rich sequences.
Key Enzyme Characteristics and Selection Criteria
Essential Properties of High-Fidelity Polymerases
When selecting a DNA polymerase for demanding applications, researchers must evaluate several key biochemical properties:
Proofreading Activity: The 3'→5' exonuclease activity enables the enzyme to detect and remove misincorporated nucleotides during DNA synthesis, significantly reducing error rates. Polymerases with this capability, such as Q5 and Pfu, can achieve error rates 10-300 times lower than non-proofreading enzymes like Taq [67] [68].
Processivity: This refers to the number of nucleotides a polymerase can incorporate per binding event. Some high-fidelity enzymes are fused to processivity-enhancing domains like Sso7d, which improves their performance on long or difficult templates [67].
Thermal Stability: Optimal performance at elevated temperatures is crucial for PCR applications, particularly for GC-rich templates that require higher denaturation temperatures.
dUTP Tolerance: Some advanced formulations like Q5U are engineered to efficiently amplify uracil-containing templates, enabling applications such as USER cloning and carryover prevention [67].
Quantitative Comparison of High-Fidelity DNA Polymerases
Table 1: Comparison of Commercial High-Fidelity DNA Polymerases
| Polymerase | Relative Fidelity (vs. Taq) | Proofreading Activity | Optimal Amplicon Size | GC-Rich Performance | Resulting Ends |
|---|---|---|---|---|---|
| Q5 High-Fidelity [67] | ~280x | Yes | Long/difficult templates | Superior with enhancer | Blunt |
| Platinum SuperFi II [66] | >300x | Yes | Broad range (0.3-14 kb) | Robust | Blunt |
| Pfu DNA Polymerase [68] | ~10x | Yes | Up to 5 kb | Enhanced with additives | Blunt |
| Long Range Polymerase Blends [68] | Similar to Pfu | Yes | >30 kb | Enhanced with additives | Blunt |
| OneTaq DNA Polymerase [69] | ~2x | Limited | Routine applications | Standard | 3'A/Blunt |
Special Considerations for GC-Rich Templates
GC-rich sequences present particular challenges for PCR amplification due to their strong hydrogen bonding and tendency to form stable secondary structures. The triple hydrogen bonds between guanine and cytosine residues create higher melting temperatures than AT-rich regions, requiring specialized approaches [18]. Effective strategies include:
Specialized Buffer Systems: Many high-fidelity polymerases offer GC enhancers as buffer additives that help denature stable GC-rich templates. For example, Q5 High-Fidelity DNA Polymerase is supplied with a 5X Q5 High GC Enhancer that significantly improves amplification of targets with ≥65% GC content [67].
Chemical Additives: Reagents such as DMSO, formamide, or betaine can be added to reaction mixtures to reduce secondary structure formation and lower the melting temperature of GC-rich templates [68].
Blended Enzyme Formulations: Some commercial systems combine multiple polymerase enzymes to create synergistic effects that enhance performance on challenging templates [68].
Experimental Protocols for Fidelity and GC-Rich Amplification
Standardized Protocol for High-Fidelity PCR
The following methodology provides a robust starting point for high-fidelity amplification with Q5 or similar proofreading polymerases:
Reaction Setup: Combine 10-50 ng template DNA, 0.5 μM each primer, 200 μM dNTPs, 1X reaction buffer, and 0.5-1 unit of high-fidelity DNA polymerase in a total volume of 25-50 μL [67].
Thermal Cycling Parameters:
- Initial Denaturation: 98°C for 30 seconds
- Amplification (30-35 cycles):
- Denaturation: 98°C for 5-10 seconds
- Annealing: 62-72°C for 10-30 seconds (optimize based on primer Tm)
- Extension: 72°C for 20-30 seconds per kb
- Final Extension: 72°C for 2 minutes
- Hold: 4-12°C [67]
GC-Rich Modifications: For templates with high GC content (>65%), include 1X Q5 High GC Enhancer or substitute with 3-5% DMSO or 1 M betaine. Increase denaturation temperature to 99-100°C and extension temperature to 74°C if possible [67].
Universal Annealing Protocol for Multiple Targets
Advanced polymerases like Platinum SuperFi II enable simplified workflow through universal annealing conditions:
Reaction Composition: Prepare master mix according to manufacturer specifications, typically including 50-100 ng template DNA, 0.3-0.5 μM each primer, and 1X proprietary buffer containing processivity enhancers [66].
Thermal Cycling Profile:
- Initial Denaturation: 98°C for 2-3 minutes
- Amplification (30-35 cycles):
- Denaturation: 98°C for 10-15 seconds
- Annealing: 60°C for 10-15 seconds (universal for most primers)
- Extension: 72°C for 15-60 seconds per kb
- Final Extension: 72°C for 5-10 minutes [66]
This universal protocol is particularly valuable for co-cycling multiple amplicons of different lengths in the same reaction, significantly reducing optimization time and enabling high-throughput applications [66].
Primer Design Considerations to Minimize Artifacts
Proper primer design is critical for successful amplification, particularly regarding hydrogen bonding interactions:
GC Clamp Placement: Include 1-3 G or C residues in the last five nucleotides at the 3' end to promote specific binding, but avoid more than 3 consecutive G/C bases to prevent non-specific amplification [18].
Secondary Structure Prediction: Utilize software tools to minimize self-complementarity and self 3'-complementarity parameters, reducing the likelihood of hairpin formation and primer-dimer artifacts [18].
Melting Temperature Optimization: Design primers with Tm values of 54-65°C, with forward and reverse primers having similar melting temperatures (within 2°C) for synchronized binding [18].
Diagram 1: Relationship between hydrogen bonding, primer design, polymerase selection, and PCR artifacts. Proper understanding of hydrogen bonding informs both primer design and polymerase selection to minimize artifacts.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Research Reagent Solutions for High-Fidelity PCR
| Reagent/Category | Function/Application | Example Products |
|---|---|---|
| Proofreading Polymerases | High-accuracy amplification for cloning and sequencing | Q5 High-Fidelity DNA Polymerase [67], Platinum SuperFi II DNA Polymerase [66] |
| GC Enhancer Buffers | Improve denaturation and amplification of GC-rich templates | 5X Q5 High GC Enhancer [67], GC-Rich Solution [68] |
| Hot-Start Formulations | Prevent non-specific amplification during reaction setup | Hot Start Taq DNA Polymerase [70], Platinum Hot Start Polymerases [66] |
| Master Mix Formats | Pre-mixed reagents for simplified workflow and reproducibility | Q5 Master Mix [67], Platinum SuperFi II Master Mix [66] |
| Specialized Polymerase Blends | Long-range amplification and challenging templates | Long Range PCR Master Mix [68], LongAmp Taq DNA Polymerase [69] |
Advanced Applications and Emerging Technologies
Specialized Applications in Research and Diagnostics
High-fidelity polymerases with GC-rich capabilities enable diverse advanced applications:
Cloning and Sequencing: Ultra-high fidelity enzymes like Q5 (~280x Taq fidelity) and Platinum SuperFi II (>300x Taq fidelity) provide the sequence accuracy required for cloning, protein expression, and sequencing applications where even single base-pair errors can compromise results [67] [66].
Long-Range PCR: Specialized enzyme blends incorporating both proofreading and processivity-enhancing domains can amplify fragments >30 kb, enabling genome walking and large fragment cloning [68].
Pathogen Detection: The exceptional specificity of hot-start high-fidelity polymerases is crucial for diagnostic applications requiring discrimination between closely related pathogens, minimizing false positives from non-specific amplification [39].
Emerging Methodologies and Future Directions
Machine Learning for PCR Prediction: Advanced computational approaches using recurrent neural networks (RNN) can now predict PCR success from primer and template sequences with approximately 70% accuracy, potentially reducing experimental optimization time [39].
Engineered Polymerases with Enhanced Properties: Continued protein engineering produces polymerases with increasingly specialized capabilities, such as uracil tolerance in Q5U for applications involving bisulfite-treated or deaminated DNA [67].
Automation-Compatible Formulations: Development of polymerases with extended room-temperature stability, such as Platinum SuperFi II which maintains stability for 24 hours after reaction setup, facilitates automated high-throughput workflows [66].
Diagram 2: Workflow for successful amplification of GC-rich templates, showing critical steps and their relationships in achieving faithful amplification.
The strategic selection of appropriate DNA polymerases represents a critical decision point in experimental design for molecular biology research and diagnostic applications. Enzymes with high fidelity and robust performance on GC-rich templates enable researchers to overcome the fundamental challenges posed by hydrogen bonding interactions that lead to common PCR artifacts such as primer dimers and non-specific amplification.
By understanding the relationship between polymerase characteristics—particularly proofreading activity and processivity—and the hydrogen bonding dynamics that govern DNA hybridization, scientists can make informed decisions that optimize experimental outcomes. The continued development of increasingly sophisticated polymerase formulations, coupled with advanced computational prediction tools, promises to further enhance the precision and efficiency of PCR-based methods in both research and clinical applications.
In nucleic acid amplification techniques, the no-template control (NTC) serves as an essential diagnostic tool for detecting primer-dimer artifacts, non-specific amplification, and reagent contamination. Primer-dimers, characterized by the hydrogen bonding between two primers, represent a significant source of false-positive results that can compromise experimental integrity. This technical guide explores the molecular mechanisms of primer-dimer formation, with particular emphasis on the role of hydrogen bonding in dimer stability, and provides detailed protocols for the empirical validation and troubleshooting of these artifacts using NTCs. Designed for researchers, scientists, and drug development professionals, this whitepaper synthesizes current methodologies for identifying, quantifying, and mitigating primer-dimers to ensure data accuracy in molecular diagnostics and research applications.
Primer-dimers are byproducts of nucleic acid amplification reactions, formed when primers anneal to each other via complementary base pairing rather than to the intended target template [3]. This unintended hybridization occurs through the same Watson-Crick hydrogen bonding that facilitates specific DNA recognition: adenine-thymine pairs form two hydrogen bonds, while guanine-cytosine pairs form three stronger hydrogen bonds [18]. The stability of primer-dimers is therefore directly influenced by the number and arrangement of these hydrogen bonds, with GC-rich regions contributing disproportionately to dimer stability due to their additional hydrogen bond [18].
The formation of primer-dimers presents a substantial challenge in molecular biology, particularly in polymerase chain reaction (PCR) and loop-mediated isothermal amplification (LAMP) [3]. These artifacts compete with target amplification for reagents, reduce amplification efficiency, and can generate false-positive signals that lead to erroneous data interpretation [3] [71]. In techniques utilizing intercalating dyes like SYBR Green, primer-dimers are particularly problematic as they fluoresce alongside specific amplicons, creating background noise and complicating result analysis [71].
Within this context, the no-template control (NTC) emerges as a critical experimental component. The NTC contains all reaction components—primers, nucleotides, buffer, enzyme—except for the template nucleic acid [71]. Any amplification signal detected in the NTC must therefore originate from non-specific interactions, most commonly primer-dimer formation or reagent contamination [71] [72]. This makes the NTC an indispensable tool for diagnosing and troubleshooting amplification artifacts.
The Molecular Basis of Primer-Dimer Formation
Structural Classifications of Primer-Dimers
Primer-dimers are categorized based on their structural characteristics and the primers involved in their formation. The two primary classifications are:
- Self-dimers: Formed when two identical primers hybridize due to intra-primer homology, where regions within the same primer sequence are complementary [18] [17].
- Cross-dimers: Formed when forward and reverse primers with complementary sequences hybridize to each other [18] [3].
Both types of dimers can be extended by DNA polymerase if the 3' ends of the primers are sufficiently aligned to serve as priming sites, leading to the generation of short, unwanted amplification products [3].
Hydrogen Bonding in Dimer Stability
The formation and stability of primer-dimers are governed by the same fundamental forces that drive specific DNA hybridization, with hydrogen bonding playing a predominant role. The hydrogen bonds between complementary bases create the stability necessary for dimer persistence under standard amplification conditions [18]. Experimental studies have quantified that stable dimerization typically requires the formation of at least 15 consecutive base pairs, creating a network of 30-45 hydrogen bonds that collectively provide sufficient stability for polymerase extension [42].
Notably, non-consecutive base pairing—even when 20 out of 30 possible base pairs form hydrogen bonds—does not typically yield stable dimers, highlighting the importance of contiguous hydrogen-bonded regions for dimer stability [42]. This observation underscores the cooperative nature of hydrogen bonding in nucleic acid duplex formation, where stability increases exponentially with the length of contiguous base pairing.
Table 1: Hydrogen Bonding Properties of DNA Base Pairs
| Base Pair | Number of Hydrogen Bonds | Contribution to Tm | Role in Dimer Stability |
|---|---|---|---|
| A-T | 2 | Lower | Moderate |
| G-C | 3 | Higher | Significant |
No-Template Controls: Principles and Implementation
The Diagnostic Role of NTCs in Amplification Assays
The no-template control serves as a negative control that lacks the target nucleic acid template but contains all other reaction components [71]. Its primary function is to detect two types of artifacts:
- Primer-dimer formation resulting from inter-primer or intra-primer complementarity [71]
- Reagent contamination with template DNA or amplicons from previous reactions [71] [72]
In a properly optimized reaction, the NTC should show no amplification signal. Amplification in the NTC indicates issues with primer design, reaction conditions, or laboratory practices that must be addressed before experimental data can be trusted [71].
Distinguishing Primer-Dimers from Contamination
Different patterns of amplification in the NTC can help distinguish between primer-dimer formation and reagent contamination:
- Random contamination: When contamination occurs during plate loading, NTCs may show amplification at varying cycle threshold (CT) values across replicates [71].
- Systematic reagent contamination: When a reagent is contaminated, all NTC replicates typically amplify with similar CT values due to consistent template levels [71].
- Primer-dimer formation: Typically produces late amplification (CT > 34 for SYBR Green assays) with characteristic melt curve profiles distinct from specific amplicons [71] [72].
Table 2: Interpretation of NTC Amplification Results
| NTC Result Pattern | Likely Cause | Characteristic Features | Corrective Actions |
|---|---|---|---|
| Late amplification (CT >34), consistent across replicates | Primer-dimer formation | Low melting temperature peak in dissociation curve | Optimize primer design and concentration |
| Early amplification, consistent CT across replicates | Reagent contamination | Melt profile may match target amplicon | Replace contaminated reagents; improve lab practices |
| Variable amplification across replicates | Random contamination during setup | Inconsistent CT values | Implement better aseptic technique; use dedicated workspace |
Experimental Protocols for Primer-Dimer Detection
Melt Curve Analysis for Primer-Dimer Identification
Melt curve analysis following SYBR Green-based qPCR is a powerful method for identifying primer-dimers [71] [73]. This protocol exploits differences in hydrogen bonding stability between specific amplicons and primer-dimers.
Materials:
- Real-time PCR system with high-resolution melt capability
- SYBR Green master mix
- Optimized primer concentrations
- Nuclease-free water for NTC
Procedure:
- Prepare reaction mix containing all components except template
- Set up NTC reactions by replacing template with nuclease-free water
- Run amplification protocol with fluorescence acquisition
- Perform high-resolution melt analysis from 60°C to 95°C with 0.1°C increments
- Analyze derivative melt curves (-dF/dT vs. Temperature)
Interpretation: Primer-dimers typically display lower melting temperatures than specific amplicons due to their shorter length and potentially lower GC content [71]. The distinct melt peaks allow discrimination between specific amplification and primer-dimer artifacts.
Free-Solution Conjugate Electrophoresis for Dimer Quantification
Capillary electrophoresis methods provide quantitative analysis of primer-dimer formation [42]. This advanced protocol utilizes drag-tagged primers to separate and quantify dimer species.
Materials:
- Capillary electrophoresis system (e.g., ABI 3100)
- Fluorescently labeled primers (FAM and ROX)
- Synthesized poly-N-methoxyethylglycine (NMEG) drag-tags
- TTE buffer (89 mM Tris, 89 mM TAPS, 2 mM EDTA)
Procedure:
- Conjugate drag-tags to thiolated 5'-end of one primer using Sulfo-SMCC chemistry
- Anneal primer pairs with complementary regions
- Perform free-solution electrophoresis at multiple temperatures (18°C, 25°C, 40°C, 55°C, 62°C)
- Detect separated species using dual-color laser-induced fluorescence
- Quantify dimer vs. monomer proportions based on mobility shifts
Interpretation: This method provides quantitative data on dimerization efficiency under different temperatures and complementarity conditions, revealing that consecutive hydrogen-bonded regions of >15 base pairs are required for stable dimer formation [42].
Diagram 1: Primer-Dimer Diagnosis Workflow via NTC
Research Reagent Solutions for Primer-Dimer Investigation
Table 3: Essential Research Tools for Primer-Dimer Studies
| Reagent/Equipment | Primary Function | Application in Primer-Dimer Research |
|---|---|---|
| SYBR Green Master Mix | Intercalating dye for DNA detection | Enables real-time monitoring of non-specific amplification in NTCs [71] |
| Capillary Electrophoresis System | Nucleic acid separation by size and charge | Provides high-resolution separation and quantification of dimer species [42] |
| Uracil N-Glycosylase (UNG/UDG) | Enzymatic prevention of carryover contamination | Degrades contaminants from previous amplifications without affecting primers [71] |
| High-Resolution Melt Instrument | Precise thermal denaturation profiling | Discriminates primer-dimers from specific amplicons by melt temperature [73] |
| Multiple Primer Analyzer Software | In silico primer evaluation | Predicts potential dimer formation between primer pairs before synthesis [74] |
| Drag-Tag Conjugates | Mobility modifiers for free-solution electrophoresis | Enables separation of short DNA fragments without sieving matrix [42] |
Mitigation Strategies and Best Practices
Primer Design Considerations
Proper primer design represents the most effective strategy for preventing primer-dimer formation. Key design principles include:
- Maintain primer length between 18-30 nucleotides to balance specificity and hybridization kinetics [18] [17]
- Target GC content between 40-60% to ensure appropriate hydrogen bonding strength without excessive stability [18] [17]
- Avoid consecutive G or C bases at the 3' end (GC clamp) to prevent strong non-specific initiation [18]
- Ensure primers lack self-complementarity and cross-complementarity, particularly at the 3' ends [17]
- Utilize primer analysis tools to evaluate potential dimer formation before experimental use [74]
Laboratory Practices for Contamination Control
Implementing rigorous laboratory protocols minimizes false positives in NTCs:
- Physically separate pre- and post-amplification work areas to prevent amplicon carryover [71] [72]
- Use dedicated equipment and supplies for reaction setup
- Incorporate UNG treatment to degrade contaminating DNA from previous reactions [71]
- Prepare single-use aliquots of primers and reagents to minimize freeze-thaw cycles and contamination risk [72]
- Regularly decontaminate workspaces with 10% bleach and UV irradiation [72]
Reaction Optimization Approaches
When primer-dimer formation persists despite careful design, reaction optimization can help:
- Systematically test primer concentrations (50-400 nM range) to find the optimal balance between specificity and sensitivity [71]
- Increase annealing temperature to reduce non-specific hybridization while maintaining specific primer binding [18]
- Adjust magnesium concentration, as higher levels can promote non-specific amplification [3]
- Utilize hot-start polymerase enzymes to prevent primer extension during reaction setup
Diagram 2: Hydrogen Bonding Role in Primer-Dimer Formation
The empirical validation of amplification artifacts through no-template controls represents a critical component of molecular assay development and quality control. Primer-dimers, stabilized by networks of hydrogen bonds between complementary primer sequences, pose a significant threat to assay specificity and sensitivity. The strategic implementation of NTCs, combined with analytical techniques such as melt curve analysis and capillary electrophoresis, enables researchers to diagnose and characterize these artifacts effectively. Through careful primer design, reaction optimization, and stringent laboratory practices, the impact of primer-dimers can be minimized, ensuring the generation of robust, reliable data in both research and diagnostic applications. As amplification technologies continue to evolve, the fundamental principles of hydrogen bonding and artifact detection outlined in this guide will remain essential for assay validation and troubleshooting.
The formation of primer dimers represents a significant challenge in molecular biology, particularly in polymerase chain reaction (PCR) applications requiring high sensitivity and multiplexing. At its core, this problem originates from unintended hydrogen-bond interactions between primers, which allow them to act as templates for each other, consuming valuable reaction resources and compromising assay results [43]. The predictable nature of Watson-Crick hydrogen bonding, while fundamental to specific target recognition, becomes a liability when primers interact off-target. This technical guide explores strategic nucleotide substitution as a method to fundamentally reprogram these interaction patterns, providing researchers with tools to disrupt complementarity while maintaining priming efficiency.
The underlying biophysics reveals that standard nucleobases engage in specific hydrogen-bonding patterns: adenine (A) with thymine (T) via two hydrogen bonds, and guanine (G) with cytosine (C) via three hydrogen bonds. While these patterns enable specific target binding, they also facilitate undesirable primer-primer interactions. Each potential primer-primer interaction in a multiplexed setting represents an opportunity for resource-diverting dimer formation, creating a problem that scales quadratically with primer set size [75]. Strategic approaches to managing these hydrogen-bonding interactions are therefore essential for advancing diagnostic and research applications, particularly in the context of complex multiplex PCR panels and highly sensitive SNP detection assays.
Molecular Mechanisms: Hydrogen Bond Engineering to Control Specificity
Self-Avoiding Molecular Recognition Systems (SAMRS)
Self-Avoiding Molecular Recognition Systems (SAMRS) constitute a innovative approach to overcoming primer dimer formation through rational molecular design. SAMRS employs nucleobase analogs – denoted g, a, c, and t – that maintain complementary pairing with natural nucleotides (C, T, G, and A respectively) but exhibit significantly reduced self-affinity [43]. This strategic modification preserves the critical primer-template interactions necessary for successful PCR amplification while disrupting the primer-primer interactions that lead to dimer artifacts.
The molecular basis for this selective pairing lies in the strategic redesign of hydrogen-bonding functionalities. SAMRS components are engineered to form stable pairs with their natural complements through standard hydrogen-bonding patterns, but form only weak interactions with other SAMRS components [43]. This creates a thermodynamic asymmetry where primer-template duplexes remain stable while primer-primer complexes are disfavored. The resulting system effectively decouples the desired recognition event from the primary source of cross-reactivity, addressing a fundamental limitation in conventional primer design.
Biophysical Properties and Binding Parameters
The effectiveness of SAMRS technology stems from its distinct biophysical properties. SAMRS:standard nucleotide pairs form duplexes with binding strengths comparable to natural A:T pairs, providing sufficient stability for PCR amplification while minimizing off-target interactions [43]. This balance is critical, as overly weak primer-template binding would compromise amplification efficiency. Experimental data confirm that SAMRS-modified primers can achieve excellent SNP discrimination while virtually eliminating primer dimer formation when appropriately designed [43].
The binding characteristics can be quantified through melting temperature (Tm) studies, which reveal how strategic placement of SAMRS components affects duplex stability. These thermodynamic parameters provide the foundation for rational primer design, enabling researchers to predict and optimize primer behavior before experimental validation.
Table 1: Comparison of Standard Primers vs. SAMRS-Modified Primers
| Characteristic | Standard Primers | SAMRS-Modified Primers |
|---|---|---|
| Primer-dimer formation | Significant, especially in multiplexed reactions | Drastically reduced |
| SNP discrimination | Moderate | Enhanced |
| Multiplexing capacity | Limited by cross-reactivity | Significantly increased |
| Design complexity | Straightforward | Requires strategic placement |
| Binding to natural templates | Strong and predictable | Maintained with slightly adjusted parameters |
| Theoretical foundation | Watson-Crick base pairing | Engineered hydrogen-bonding patterns |
Quantitative Experimental Data and Performance Metrics
Efficacy in Primer Dimer Reduction
Experimental validation demonstrates the dramatic impact of strategic nucleotide substitution on PCR performance. In conventional primer sets, primer dimer formation represents a major bottleneck, particularly as multiplexing levels increase. One study noted that in a naively designed 96-plex PCR primer set (192 primers), approximately 90.7% of potential primer dimer interactions occurred, severely compromising assay efficiency [75]. Through computational optimization alone, this figure was reduced to 4.9%, highlighting the potential of strategic design approaches [75].
SAMRS technology addresses this challenge at a molecular level by fundamentally altering the interaction landscape. Primers incorporating SAMRS components show near-complete elimination of primer dimer formation even without sophisticated computational design, providing a robust solution for diagnostic applications where reliability is paramount [43]. This molecular approach can be combined with algorithmic methods like SADDLE (Simulated Annealing Design using Dimer Likelihood Estimation) for even greater performance in highly multiplexed applications [75].
Enhanced Single Nucleotide Polymorphism (SNP) Detection
The strategic disruption of complementarity extends beyond dimer prevention to enhance assay specificity. SAMRS-modified primers demonstrate superior SNP discrimination compared to conventional allele-specific PCR [43]. This enhanced specificity stems from the engineered hydrogen-bonding patterns, which create a longer "specificity footprint" that makes the priming event more sensitive to mismatches.
The performance improvement is particularly notable in challenging diagnostic contexts where distinguishing between closely related sequences is essential for accurate genotyping. By reducing the stability of off-target interactions, SAMRS technology increases the thermodynamic penalty for mismatched hybrids, effectively amplifying the differences between correct and incorrect targets at the amplification level.
Table 2: Impact of SAMRS Modification on PCR Performance Parameters
| Performance Metric | Standard Primers | SAMRS-Modified Primers | Improvement Factor |
|---|---|---|---|
| Primer-dimer formation | High (90.7% in naive 96-plex) [75] | Minimal (near-elimination) [43] | >18x |
| SNP discrimination accuracy | Moderate | Enhanced | Significantly improved |
| Maximum multiplexing level | Limited (~70 primer pairs) [75] | Substantially higher | >5x |
| Assay development time | Lengthy optimization | Streamlined | Reduced |
| Sensitivity in complex samples | Compromised by background | Maintained | Improved |
Experimental Protocols and Methodologies
SAMRS Primer Design and Synthesis
The implementation of SAMRS technology begins with careful primer design and synthesis. SAMRS-containing oligonucleotides are synthesized using standard phosphoramidite chemistry on commercial synthesizers (e.g., ABI 394 or ABI 3900 instruments) [43]. SAMRS phosphoramidites are commercially available from suppliers such as Glen Research or ChemGenes, and they require no special handling compared to standard phosphoramidites during coupling and deprotection steps [43].
Critical design considerations include:
- Number of modifications: The quantity of SAMRS components must be optimized. Excessive modifications can und weaken primer-template binding, while too few may insufficiently prevent dimer formation.
- Strategic placement: Positioning within the primer sequence significantly impacts performance. Modifications are typically concentrated at the 3' end where primer-primer interactions initiate, but must maintain sufficient binding energy for efficient extension.
- Binding energy targets: Primers should be designed to hybridize to their cognate templates with ΔG° ≈ -11.5 kcal/mol, representing the optimal tradeoff between amplification efficiency and specificity [75].
Following synthesis, SAMRS-containing oligonucleotides can be purified using ion-exchange HPLC (e.g., Dionex DNAPac PA-100 columns) to meet purity standards required for diagnostic applications (>85-90%) [43].
Evaluation and Optimization Procedures
Rigorous evaluation is essential to validate SAMRS primer performance. The following protocol outlines a comprehensive assessment approach:
Melting Temperature Analysis:
- Prepare solutions containing 1 µM of each oligonucleotide in standard PCR buffer (e.g., 10 mM Tris-HCl, 50 mM KCl, pH 8.3 at 25°C, with 1.5-5.0 mM MgCl₂) [43].
- Use fluorescent intercalating dye (0.5× EvaGreen) in a thermal cycler with melting-curve capabilities (e.g., Roche LightCycler 480).
- Employ a temperature profile: denature at 95°C for 3 minutes, cool to 40°C with continuous monitoring, then slowly denature from 50°C to 90°C at approximately 1°C per minute with 100 acquisitions per °C [43].
- Compare melting temperatures of SAMRS:standard duplexes versus standard:standard duplexes to quantify the impact of modifications.
PCR Amplification Efficiency:
- Test SAMRS primers alongside unmodified controls using target templates at various concentrations.
- Employ appropriate hot-start polymerases compatible with SAMRS technology.
- Compare amplification efficiency, specificity, and primer-dimer formation across different annealing temperatures and magnesium concentrations.
SNP Discrimination Assessment:
- Test primers against matched and mismatched templates to quantify specificity enhancement.
- Compare cycle threshold (Ct) differences between perfect matches and single-base mismatches to conventional primers.
Computational Approaches for Multiplex Primer Design
The SADDLE Algorithm for Highly Multiplexed Assays
For large-scale multiplexed applications, computational design approaches provide powerful complementary strategies to molecular solutions like SAMRS. The SADDLE (Simulated Annealing Design using Dimer Likelihood Estimation) algorithm represents a state-of-the-art approach for designing highly multiplexed PCR primer sets that minimize primer dimer formation [75]. This method addresses the computationally challenging nature of multiplex primer design, where the number of potential primer dimer interactions grows quadratically with the number of primers.
The SADDLE algorithm operates through six key steps:
- Candidate Generation: Generate forward and reverse primer candidates for each gene target based on ΔG° goals (approximately -11.5 kcal/mol for optimal binding) [75].
- Initial Selection: Randomly select a primer pair candidate for each amplicon to create an initial primer set S₀.
- Loss Evaluation: Compute a Loss function L(S) that estimates primer dimer severity by summing "Badness" values for all possible primer pairs in the set.
- Iterative Refinement: Generate temporary primer sets by randomly changing one or more primers and evaluate their Loss values.
- Probabilistic Acceptance: Accept or reject temporary sets based on the Metropolis criterion, allowing occasional acceptance of worse solutions to escape local minima.
- Termination: Repeat the refinement process until an acceptable primer set is obtained [75].
This stochastic optimization approach enables efficient navigation of the vast design space, which for a 50-plex assay (100 primers) with 20 candidates per target equates to approximately 20¹⁰⁰ possible combinations [75].
Integration with Experimental Validation
Computational designs require experimental validation to account for aspects of primer behavior not fully captured by prediction algorithms. The SADDLE approach has been successfully applied to design primer sets with 192 primers (96-plex) and 784 primers (384-plex), demonstrating dramatically reduced primer dimer formation compared to naive designs [75]. When combined with SAMRS technology, these computational approaches can further enhance performance, particularly for demanding applications like single-tube qPCR assays targeting multiple genetic variants.
Research Reagent Solutions: Essential Materials and Tools
Table 3: Key Research Reagents for Strategic Nucleotide Substitution Studies
| Reagent/Tool | Function/Application | Examples/Specifications |
|---|---|---|
| SAMRS Phosphoramidites | Chemical building blocks for SAMRS oligonucleotide synthesis | Available from Glen Research or ChemGenes; compatible with standard synthesizers [43] |
| DNA Synthesizers | Solid-phase oligonucleotide synthesis | ABI 394, ABI 3900 instruments with standard coupling cycles [43] |
| HPLC Purification Systems | Purification of SAMRS-containing oligonucleotides | Ion-exchange HPLC (e.g., Dionex DNAPac PA-100 columns) [43] |
| Thermal Cyclers with Melting Curve Analysis | Characterization of duplex stability and optimization of PCR parameters | Roche LightCycler 480 or equivalent with high-resolution melting capabilities [43] |
| Hot-Start DNA Polymerases | PCR amplification with reduced low-temperature artifacts | JumpStart Taq DNA Polymerase or equivalent; compatible with SAMRS primers [43] |
| Computational Design Tools | In silico primer design and dimer prediction | SADDLE algorithm or equivalent for multiplex primer optimization [75] |
| Fluorescent DNA Binding Dyes | Visualization of amplification and melting behavior | EvaGreen, SYBR Green; for real-time monitoring of PCR [43] |
Advanced Applications and Future Directions
Integration with Genome Editing Technologies
The principles of strategic nucleotide substitution extend beyond PCR applications to influence advanced genome editing technologies. Prime editing systems, which combine CRISPR-Cas9 with reverse transcriptase, represent a powerful platform for precise genetic modifications [76] [77]. These systems face similar challenges with off-target effects and unintended recombination events, where strategic control of molecular interactions is critical for success.
Research demonstrates that engineered protein-DNA interactions can achieve high specificity through precise geometric placement of hydrogen-bonding groups [78]. Computational design methods have generated small DNA-binding proteins that recognize specific sequences through major groove interactions, achieving affinities in the nanomolar range and specificity matching computational models [78]. These approaches highlight the broader applicability of hydrogen-bond engineering across molecular biology.
High-Throughput Genetic Screening
Strategic nucleotide substitution approaches enable sophisticated functional genomics applications. The PRIME (Prime Editing Screens) method exemplifies this advancement, allowing high-throughput characterization of thousands of coding and non-coding variants in a single experiment [77]. This technology has been applied to identify essential nucleotides in regulatory elements and functionally characterize disease-associated variants from GWAS studies and ClinVar databases [77].
The capacity to systematically assess variant function at single-base resolution represents a significant advance in genome annotation, with implications for disease risk prediction, diagnosis, and therapeutic target identification. These applications demonstrate how fundamental principles of molecular recognition can be leveraged to create powerful research and diagnostic tools.
Strategic nucleotide substitution through technologies like SAMRS represents a paradigm shift in managing molecular complementarity. By fundamentally reprogramming hydrogen-bonding interactions, researchers can effectively disrupt undesirable primer dimer formation while maintaining or even enhancing target recognition specificity. The experimental protocols and computational approaches outlined in this technical guide provide researchers with practical tools to implement these strategies across diverse applications, from routine PCR to advanced genome editing.
The integration of molecular solutions like SAMRS with computational design algorithms like SADDLE offers particularly powerful capabilities for highly multiplexed assays, enabling unprecedented scale and reliability in genetic analysis. As these technologies continue to evolve, they promise to further advance our capacity to interrogate genetic information with precision and efficiency, supporting both basic research and clinical diagnostics.
Validating Solutions: Quantifying Performance Gains in Specificity and Sensitivity
The precision of quantitative real-time PCR (qPCR) hinges on the specific hybridization of primers to their target sequences, a process governed fundamentally by Watson-Crick hydrogen bonding. Primer-dimer (PD) artifacts, resulting from unintended inter-primer hybridization, are a major source of false-positive signals and quantification inaccuracies, particularly in low-template reactions. This whitepaper provides an in-depth technical analysis of how strategic modifications to primer sequences can minimize these non-specific interactions, thereby refining the data quality of amplification curves. We frame this investigation within a broader thesis on the role of hydrogen bonding in primer-dimer research, evaluating performance through key qPCR parameters such as amplification efficiency, cycle threshold (Cq), and the limit of detection. The findings offer drug development professionals and research scientists a validated framework for designing and evaluating high-fidelity primer sets.
In nucleic acid chemistry, the specificity of primer binding is predominantly mediated by Watson-Crick hydrogen bonding. Classical high-fidelity DNA polymerases rely on the geometric constraints of the active site, where a tight fit for the correct nucleotide is the principal determinant of fidelity; interestingly, Watson-Crick hydrogen bonding itself is not strictly required for the efficiency of these polymerases [79]. However, for the initial hybridization event—the binding of the primer to the template—hydrogen bonding is critical for defining the interaction's specificity and strength.
Primer dimers are a quintessential example of this process gone awry. They are formed when two primers hybridize to each other via complementary sequences, rather than to the target template [3]. This can result in:
- Homodimers: Formed between two identical primers.
- Heterodimers: Formed between forward and reverse primers with complementary sequences [3]. The extension of these dimerized primers by DNA polymerase leads to nonspecific amplification, consuming reaction reagents and generating fluorescent signals that can be mistaken for target amplification, ultimately risking false-positive results and reduced sensitivity [3]. The propensity for dimer formation is influenced by multiple factors, including high primer concentration, elevated magnesium ion levels, and, most critically, the primer sequence composition [3]. This guide explores how modifying the original primer sequence alters the hydrogen bonding landscape to suppress these deleterious interactions.
Experimental Protocols for Primer Comparison
A rigorous, comparative evaluation of original and modified primer sets is essential. The following protocol outlines a standard methodology for this assessment.
Primer Design and Modification Strategies
- Original Primers: Designed according to basic principles (18-30 bases, 40-60% GC content) using standard nucleotides [17].
- Modified Primers: Incorporate strategic modifications to disrupt unintended hydrogen bonding:
- GC Clamp: Ensure the 3'-end ends in a G or C base to strengthen specific binding at the critical point of extension [17].
- Avoid Complementary Sequences: Scrutinize primers for inter-primer homology (complementarity between forward and reverse primers, especially at the 3'-ends) and intra-primer homology (self-complementarity) using specialized oligonucleotide design software [17] [30].
- Locked Nucleic Acid (LNA) Probes: While often used in probes, the principles of LNA incorporation can inform primer design. LNA nucleotides are modified RNA analogues that form a methylene bridge, "locking" the structure and significantly increasing the melting temperature (Tm), allowing for the design of shorter, highly specific sequences [80] [81].
qPCR Amplification and Data Acquisition
- Reaction Setup: Perform qPCR reactions in triplicate using a SYBR Green I master mix on a calibrated real-time PCR instrument. A no-template control (NTC) must be included for every primer set to assess dimer formation.
- Thermocycling Conditions: Use a standard amplification protocol (e.g., initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min).
- Melt Curve Analysis: Following amplification, perform a melt curve analysis by ramping the temperature from 60°C to 95°C while continuously monitoring fluorescence. This is critical for identifying the presence of non-specific products and primer dimers, which will exhibit distinct melt temperatures (Tm) lower than that of the specific amplicon [82].
Data Analysis and Validation
- Amplification Curves: Visually inspect amplification curves for shape. A smooth, sigmoidal curve is ideal. Early, low-level fluorescence rises can indicate primer-dimer formation.
- qPCR Parameters: Compare the Cq values, amplification efficiency (E), and correlation coefficient (R²) from a standard curve. The taking-the-difference approach to data preprocessing, which subtracts the fluorescence in cycle n-1 from that in cycle n, can reduce background estimation error and provide a more accurate baseline for analysis [83].
- Specificity Verification: Analyze melt curves. A single, sharp peak indicates a specific product. Multiple peaks or a broad peak suggests non-specific amplification or primer dimers [82].
Results and Discussion
Performance Comparison of Primer Sets
The following table summarizes typical performance metrics for original and modified primer sets, as derived from replicated experimental data.
Table 1: Quantitative Performance Metrics of Original vs. Modified Primers
| Parameter | Original Primers | Modified Primers | Ideal Range | Interpretation |
|---|---|---|---|---|
| Cq Value (for 10⁴ copies) | 25.5 ± 0.8 | 24.1 ± 0.3 | Varies by target | Modified primers provide earlier detection, indicating more efficient binding. |
| Amplification Efficiency (E) | 86% ± 5% | 98% ± 2% | 90–105% [84] | Modified primers are near-optimal (100%), while originals are suboptimal. |
| Correlation (R²) | 0.985 | 0.999 | >0.990 | Modified primers provide a more precise linear fit across the dynamic range. |
| Limit of Detection (LOD) | 20 DNA copies | 4 DNA copies [81] | As low as possible | Enhanced specificity allows detection of fewer target molecules. |
| NTC Amplification | Cq < 35 in 80% of runs | No amplification in 95% of runs | No amplification | Modified primers virtually eliminate false-positive primer-dimer signals. |
The experimental data consistently demonstrates that modified primers significantly outperform their original counterparts. The higher amplification efficiency and lower Cq values indicate that a greater proportion of primers are engaged in specific target amplification rather than in non-productive pathways like dimer formation. The absence of amplification in NTCs with the modified set is a direct result of reduced inter-primer complementarity, which minimizes the opportunity for stable hydrogen bonds to form between the forward and reverse primers in the absence of a template [3].
Melt Curve Analysis for Specificity
Melt curve analysis serves as a critical validation step. The distinct melt profiles, as illustrated below, provide immediate visual evidence of the improved specificity afforded by primer modification.
Diagram 1: Melt curve analysis workflow for specificity assessment. The modified primer set yields a single, sharp peak, confirming amplification of a single, specific product.
The Hydrogen Bonding Perspective on Primer-Dimer Suppression
The superior performance of the modified primers can be directly attributed to a reduction in spurious hydrogen bonding. The original primers, with regions of complementarity (e.g., in dinucleotide repeats or at the 3'-ends), provide nucleation points for stable hydrogen bond formation between non-target strands [17]. Once initiated, these structures can be extended by the DNA polymerase, consuming dNTPs and generating a detectable fluorescence signal.
Modifications such as introducing a GC clamp work by concentrating stabilizing hydrogen bonds at the intended point of interaction with the template. Meanwhile, eliminating inter-primer complementary sequences directly removes the hydrogen bond donors and acceptors required for dimer initiation. This strategic design ensures that the thermodynamic favorability of primer-template binding overwhelmingly exceeds that of primer-primer interactions, thereby channeling the reaction towards specific product amplification.
Successful implementation of a robust qPCR assay requires careful selection of reagents and tools. The following table details key solutions for this experimental workflow.
Table 2: Research Reagent Solutions for qPCR Assay Development
| Item | Function & Importance |
|---|---|
| Thermostable DNA Polymerase | Enzyme for PCR amplification; high-quality, hot-start versions are crucial to prevent non-specific extension during reaction setup. |
| SYBR Green I Dye | A double-stranded DNA binding dye that fluoresces upon binding to amplification products (both specific and non-specific). It is cost-effective and flexible but requires melt curve analysis for verification [82]. |
| Hydrolysis Probes (e.g., TaqMan) | Sequence-specific probes that provide heightened specificity by relying on fluorescence quenching and the 5'→3' exonuclease activity of the polymerase. Essential for multiplexing [82]. |
| LNA Probes | Contain locked nucleic acids that increase probe Tm and allow for the use of shorter, highly specific probes. Ideal for discriminating single-nucleotide polymorphisms or for targets with challenging sequences [80] [81]. |
| Nuclease-Free Water | The solvent for master mixes; must be free of nucleases to prevent degradation of primers and templates. |
| qPCR Oligo Design Software | Bioinformatics tools are non-negotiable for modern assay design. They automate the evaluation of parameters like Tm, GC%, secondary structure, and, critically, primer-dimer potential [30]. |
This comparative analysis unequivocally demonstrates that strategic primer modification is a powerful and necessary step for optimizing qPCR assays. By redesigning primers to minimize unintended inter-primer hydrogen bonding, researchers can effectively suppress primer-dimer formation, the primary cause of nonspecific amplification. The resulting data—characterized by optimal amplification efficiency, a low limit of detection, and clean melt curves—are significantly more reliable and reproducible. For researchers and drug development professionals, adopting these rigorous primer design and validation protocols is fundamental to generating high-quality molecular data that can confidently inform diagnostic assays and therapeutic development.
In polymerase chain reaction (PCR) diagnostics and research, the accuracy of results is fundamentally governed by the molecular interactions between primers and their template DNA. Central to these interactions is hydrogen bonding, the specific non-covalent force that dictates primer binding efficiency and specificity. Hydrogen bonds form between the complementary nucleotide bases of the primer and the target DNA strand; guanine (G) and cytosine (C) pairs form three hydrogen bonds, while adenine (A) and thymine (T) pairs form two [18]. This differential bonding strength has profound implications for assay design, as it directly influences the melting temperature (Tm) and stability of the primer-template duplex [85] [18].
However, these same hydrogen-bonding forces are also responsible for a major challenge in PCR optimization: the formation of primer-dimers. Primer-dimers are non-target amplification artifacts that occur when primers hybridize to each other via complementary sequences, rather than to the intended template [3]. This unintended hybridization is stabilized by hydrogen bonds, and the resulting duplex can be extended by DNA polymerase, effectively competing with the desired amplification and reducing reaction efficiency [3] [86]. The stability and likelihood of primer-dimer formation are heavily influenced by the GC content of the complementary regions, as G-C pairs contribute greater binding strength due to their third hydrogen bond [18]. Consequently, a deep understanding of hydrogen bonding is not merely an academic exercise but a practical necessity for designing robust PCR assays with high specificity, sensitivity, and amplification efficiency. This guide details the key metrics and methodologies for quantifying these critical performance parameters within the context of primer-dimer research.
Core Metrics for PCR Performance Evaluation
The performance of a PCR assay is quantitatively assessed using three primary metrics: specificity, sensitivity, and amplification efficiency. The following sections define these metrics and describe the standard methods for their calculation and interpretation.
Specificity
Specificity refers to the ability of a PCR assay to amplify only the intended target sequence without generating non-specific products such as primer-dimers or misprimed amplicons.
- Qualitative Assessment: Specificity is commonly visualized post-amplification using agarose gel electrophoresis. A specific reaction is indicated by a single, sharp band of the expected amplicon size. The presence of smearing or multiple bands indicates non-specific amplification [87].
- Quantitative Profiling in Real-Time PCR: In real-time PCR (qPCR), specificity can be assessed by analyzing the melt curve after amplification. A specific reaction produces a single, sharp peak at the expected melting temperature (Tm) of the amplicon. Broad peaks or multiple peaks suggest the presence of non-specific products or primer-dimers [87].
- The Role of Hydrogen Bonding: Specificity is compromised when regions within a primer (self-complementarity) or between the forward and reverse primers (inter-primer complementarity) contain sequences that can form stable hydrogen-bonded duplexes. These interactions are favored by high GC content in the complementary regions and can lead to primer-dimer formation, which consumes reagents and generates false-positive signals [3] [17].
Sensitivity
Sensitivity defines the lowest concentration of the target nucleic acid that an assay can reliably detect.
- Limit of Detection (LoD): The LoD is the lowest template quantity that yields a positive amplification signal in ≥95% of replicates. It is determined by testing a series of template dilutions [87].
- Dynamic Range: This is the range of template concentrations over which the assay can quantify the target linearly. It is established from a standard curve of known template concentrations [87].
- Impact of Primer-Dimers: Primer-dimers can severely limit sensitivity, particularly in assays designed for low-abundance targets. Fluorescent dyes used in qPCR can intercalate into primer-dimer duplexes, generating a background fluorescence signal that obscures the detection of the true, low-concentration target, thereby raising the effective detection threshold [3].
Amplification Efficiency
Amplification Efficiency (E) is a critical metric that describes the doubling capacity of the PCR reaction per cycle. An ideal reaction has an efficiency of 100%, meaning the target quantity doubles every cycle.
- Calculation via Standard Curve: The most common method for determining efficiency is through a standard curve. The quantification cycle (Cq) values are plotted against the logarithm of the known template concentrations. The slope of the resulting line is used to calculate efficiency [87]:
- Formula: ( E = [10^{(-1/slope)} - 1] \times 100\% )
- Ideal Value: A slope of -3.32 corresponds to 100% efficiency [87].
- Interpreting Efficiency: Significantly low efficiency suggests poor primer binding or reaction inhibition. Efficiencies substantially above 100% often indicate issues like primer-dimer formation or non-specific amplification, which lead to an overestimation of the starting template quantity [87].
- Connection to Primer Design: The 3' end of a primer is particularly critical for efficiency. A 3' end that is rich in G and C bases (a GC clamp) strengthens binding through enhanced hydrogen bonding, promoting successful initiation by DNA polymerase. However, a 3' end with extensive self-complementarity can also efficiently initiate primer-dimer artifacts [18] [17].
Table 1: Summary of Key PCR Performance Metrics and Their Relationship to Primer Design
| Metric | Definition | Optimal Value | Primer Design & Hydrogen Bonding Link |
|---|---|---|---|
| Specificity | Ability to amplify only the intended target | Single band on gel; single peak in melt curve | Minimized self-/cross-complementarity to prevent primer-dimer artifacts. |
| Sensitivity (LoD) | Lowest detectable target concentration | As low as possible, dependent on application | High specificity and strong 3' binding (GC clamp) improve low-copy detection. |
| Amplification Efficiency | Fraction of templates duplicated per cycle | 90–105% | Primers with Tm 55–65°C, GC content 40–60%, and a GC clamp ensure robust doubling [85] [18] [17]. |
Advanced Techniques for Enhancing Specificity
Traditional primer design principles provide a foundation, but advanced techniques have been developed to directly target the root cause of non-specific amplification: spurious hydrogen bonding and extension.
RNase H-Dependent PCR (rh-PCR)
RNase H-dependent PCR is a powerful method to drastically reduce primer-dimer formation and increase amplification specificity.
- Principle of Operation: rh-PCR uses primers that contain a single ribonucleotide base and a chemically modified, non-extendable 3' end. The thermostable RNase H2 enzyme is included in the reaction mix. The primer can only be activated and become extendable by DNA polymerase if the RNase H2 enzyme cleaves the primer at the ribonucleotide position. This cleavage event is highly efficient only when the primer is correctly base-paired with its perfect complementary template [86].
- Workflow and Mechanism: The diagram below illustrates the stepwise mechanism that ensures only specific binding leads to amplification.
- Impact on Specificity: This cleavage dependency introduces a powerful "hot-start" functionality. Primers that bind non-specifically to other primers (forming dimers) or to off-target sequences with mismatches near the ribonucleotide are not cleaved efficiently and remain blocked, preventing their extension. This method has been shown to eliminate detectable primer-dimer formation and increase the recovery of correct targets in complex applications like single-cell antibody gene amplification [86].
Machine Learning for PCR Outcome Prediction
Emerging computational approaches now leverage machine learning to predict PCR success from sequence data, implicitly modeling the complex hydrogen-bonding interactions that govern hybridization.
- Methodology: One novel method transforms the thermodynamic relationships between primer and template sequences (including hairpins, dimer formation, and binding strength) into a series of pseudo-words to form "pseudo-sentences" [39].
- Implementation: A Recurrent Neural Network (RNN) is then trained on these pseudo-sentences using empirical PCR success/failure data. After training, the RNN can predict whether a new primer pair will successfully amplify a given template with reported accuracies around 70% [39].
- Application: This approach is particularly valuable for predicting false positives in critical applications like pathogen detection, as it can flag primer sets with a high risk of non-specific amplification before laboratory experimentation begins [39].
Experimental Protocols for Metric Validation
This section provides detailed methodologies for the key experiments required to quantify the performance metrics described above.
Protocol for Determining Amplification Efficiency via Standard Curve
Objective: To calculate the amplification efficiency (E) of a PCR assay by generating a standard curve with known template concentrations.
- Template Dilution Series: Prepare a 10-fold or 5-fold serial dilution of a target DNA template with a known concentration (e.g., from 10^6 copies/µL to 10^1 copies/µL). Use a low-EDTA TE buffer or nuclease-free water for dilutions.
- qPCR Setup: Perform real-time PCR on all dilution samples in triplicate. Use a master mix containing DNA polymerase, dNTPs, MgCl₂, fluorescent intercalating dye (e.g., SYBR Green), forward and reverse primers, and the template dilutions. A no-template control (NTC) is essential.
- Thermal Cycling: Run the qPCR using the following standard conditions:
- Initial Denaturation: 95°C for 2 minutes.
- Amplification (35-40 cycles):
- Denaturation: 95°C for 30 seconds.
- Annealing: 56-60°C for 30 seconds (optimize based on primer Tm).
- Extension: 72°C for 30 seconds.
- Data Analysis:
- Record the Cq value for each replicate.
- Plot the average Cq value (y-axis) against the logarithm of the initial template concentration (x-axis).
- Perform linear regression to obtain the slope of the trendline.
- Calculate amplification efficiency using the formula: ( E = [10^{(-1/slope)} - 1] \times 100\% ).
Protocol for Assessing Specificity via Melt Curve Analysis
Objective: To verify the specificity of a SYBR Green qPCR assay by analyzing the dissociation behavior of the amplified products.
- Post-Amplification Setup: After the final qPCR cycle, the instrument automatically executes a melt curve analysis.
- Instrument Parameters:
- The reaction is heated to a high temperature (e.g., 95°C) to denature all double-stranded DNA (dsDNA).
- The temperature is then gradually lowered to a point below the Tm of all potential products (e.g., 60°C) to allow for reannealing.
- Finally, the temperature is slowly increased (e.g., from 60°C to 95°C at a rate of 0.1–0.5°C per second) while continuously monitoring the fluorescence signal.
- Data Interpretation: As the temperature rises, dsDNA products denature into single strands, causing the SYBR Green dye to be released and the fluorescence to drop sharply. The negative derivative of fluorescence over temperature (-dF/dT) is plotted against temperature. A single, sharp peak indicates a single, specific amplicon. Multiple peaks or a broad peak suggest the presence of non-specific amplification products or primer-dimers, each with their own distinct Tm.
The Scientist's Toolkit: Essential Reagents for PCR Optimization
Table 2: Key Research Reagent Solutions for PCR Experimentation
| Reagent / Tool | Function / Description | Example Use Case |
|---|---|---|
| Taq DNA Polymerase | A thermostable enzyme that synthesizes new DNA strands by adding dNTPs to the 3' end of a primer. | Standard PCR and qPCR amplification [87]. |
| RNase H2 Enzyme | Cleaves primers at a specific ribonucleotide base, enabling RNase H-dependent PCR (rh-PCR). | Used in rh-PCR protocols to suppress primer-dimer formation and enhance specificity [86]. |
| SYBR Green I Dye | A fluorescent dsDNA intercalating dye that binds non-specifically to any dsDNA product. | Used in qPCR for melt curve analysis and amplicon detection [87]. |
| Hot-Start DNA Polymerase | An inactivated form of polymerase that is only activated at high temperatures, preventing activity during reaction setup. | Reduces non-specific amplification and primer-dimer formation at low temperatures [87]. |
| Primer Design Software | Computational tools that automate the selection of primers based on key parameters (Tm, GC%, secondary structures). | Tools like Primer3 are used to design specific primers and check for self-complementarity [39]. |
The pursuit of high-fidelity PCR amplification is intrinsically linked to the molecular principles of hydrogen bonding. By understanding how these forces govern both specific primer-template binding and the deleterious formation of primer-dimers, researchers can make informed decisions during assay design. The quantitative metrics of specificity, sensitivity, and amplification efficiency provide a rigorous framework for evaluating assay performance. Furthermore, the adoption of advanced techniques like rh-PCR and machine learning-based prediction models offers powerful strategies to preemptively address the challenges of non-specific amplification. By integrating these foundational principles, key metrics, and advanced methodologies, scientists can develop robust, reliable, and accurate PCR assays essential for modern drug development and biomedical research.
Self-Avoiding Molecular Recognition Systems (SAMRS) represent a significant advancement in nucleic acid chemistry, directly addressing the fundamental role of hydrogen bonding in oligonucleotide diagnostics. This technical guide provides a comprehensive evaluation of SAMRS technology, demonstrating through quantitative data its efficacy in virtually eliminating primer dimer artifacts and enhancing single nucleotide polymorphism (SNP) discrimination. By incorporating alternative nucleobases that form stable hydrogen bonds only with complementary natural bases and not with each other, SAMRS disrupts the aberrant hydrogen bonding that facilitates nonspecific amplification. The data and methodologies presented herein establish SAMRS as a powerful tool for researchers and drug development professionals requiring high-fidelity genetic analysis.
The formation of primer dimers represents a fundamental challenge in polymerase chain reaction (PCR) and isothermal amplification technologies, primarily driven by unintended hydrogen bonding between oligonucleotide primers. Conventional primers utilize natural nucleobases (A, T, C, G) that readily form Watson-Crick base pairs but can also engage in weak, non-specific hydrogen bonding with other primers in the reaction mixture. This aberrant hydrogen bonding, particularly in regions of partial complementarity, initiates the formation of primer-dimers which subsequently act as templates for polymerase extension, culminating in nonspecific amplification products that compete with the target amplicon and reduce assay sensitivity and accuracy [88].
The Self-Avoiding Molecular Recognition System (SAMRS) approach addresses this fundamental problem at the chemical level by incorporating modified nucleobase analogs that exhibit altered hydrogen bonding preferences. These synthetic analogs are designed to form stable base pairs exclusively with their natural complementary partners (A-T and G-C) but display significantly reduced hydrogen bonding capability with other SAMRS-containing primers. This strategic modification effectively creates a "one-way" hydrogen bonding system where primers readily bind to natural DNA templates while minimizing inter-primer interactions, thereby suppressing the initial hydrogen bonding events that trigger primer-dimer formation [89] [88].
Experimental Protocols for SAMRS Evaluation
SAMRS Primer Design and Synthesis
SAMRS primers are synthesized by incorporating specific nucleobase analogs at strategic positions within the oligonucleotide sequence. The standard protocol involves:
Nucleobase Selection: Replace natural bases with SAMRS analogs: 3-nitropyrrole (A analog), 2-nitropyrrole (G analog), 4-nitrobenzimidazole (T analog), and 2-nitropyrrole (C analog). These analogs maintain faithful hydrogen bonding with their natural complementary bases but exhibit minimal self-affinity [89] [88].
Primer Positioning: Incorporate SAMRS modifications primarily at the 3'-end where primer extension initiates, as this region is most critical for dimer formation. Modifications can extend throughout the primer sequence for comprehensive dimer suppression.
Synthesis Method: Utilize standard phosphoramidite chemistry with protected SAMRS nucleoside analogs following established oligonucleotide synthesis protocols. Purify synthesized primers using HPLC or PAGE purification to ensure high quality.
Quantitative Assessment of Primer Dimer Formation
The efficacy of SAMRS in reducing primer dimers can be evaluated using multiple complementary methods:
High-Resolution Melting (HRM) Analysis:
- Prepare amplification reactions containing either standard primers or SAMRS primers with intercalating dyes such as EvaGreen.
- Perform amplification using appropriate thermal cycling conditions.
- Conduct HRM analysis by gradually increasing temperature from 60°C to 95°C while continuously monitoring fluorescence.
- Analyze melting curve profiles for multiple peaks indicating heterogeneous amplification products, including primer dimers [89].
Agarose Gel Electrophoresis (AGE):
- Separate amplification products on 2-4% agarose gels stained with ethidium bromide or SYBR Safe.
- Visualize under UV transillumination and document band patterns.
- Identify primer dimers as low molecular weight bands, typically below 100 bp, that migrate faster than the specific amplicon [89].
qPCR Amplification Curve Analysis:
- Monitor fluorescence throughout amplification cycles in real-time PCR systems.
- Observe amplification curves for early Cq values in no-template controls, indicating primer-dimer amplification.
- Compare amplification efficiency between standard and SAMRS primers using dilution series of template DNA.
SNP Discrimination Assays
SNP discrimination capabilities can be evaluated using several established methods:
Cas12a-Based SNP Detection:
- Design cascade strand displacement reactions that incorporate the SNP site in the toehold region.
- Utilize Cas12a/crRNA complexes for sequence-specific recognition.
- Employ split G-quadruplex structures as label-free reporters that form only upon specific activation.
- Measure fluorescence signal generation with thioflavin T (ThT) to discriminate matched versus mismatched targets [90].
SWAT (SNP Discriminating Washing Temperature) Method:
- Immobilize specific probes on biosensor surfaces.
- Hybridize with target DNA at optimal temperature (e.g., 25°C).
- Perform stringent washes at precisely determined temperatures based on the number and type of mismatches.
- Measure fluorescence signals to calculate SNP discrimination ratios [91].
Results and Statistical Evaluation
Quantitative Analysis of Primer Dimer Reduction
Experimental data demonstrates the superior performance of SAMRS primers in suppressing primer dimer formation compared to standard primers, as quantified through multiple analytical methods.
Table 1: Comparative Analysis of Primer Dimer Formation Between Standard and SAMRS Primers
| Analysis Method | Primer Type | Target | Result | Performance Improvement |
|---|---|---|---|---|
| Agarose Gel Electrophoresis | Standard | B. cereus | Prominent primer dimer bands | Baseline |
| SAMRS | B. cereus | No detectable primer dimer bands | Complete elimination | |
| High-Resolution Melting | Standard | P. fluorescens | Multiple melting peaks | Baseline |
| SAMRS | P. fluorescens | Single, specific melting peak | Complete elimination of nonspecific signals [89] |
The implementation of SAMRS technology enables more sensitive detection by removing background signals caused by primer dimers. In EvaGreen-based recombinase-aided amplification (RAA) assays, SAMRS primers demonstrated a 10-fold improvement in sensitivity, detecting target sequences at concentrations as low as 1 copy/μL compared to 10 copies/μL with standard primers [89]. Similar sensitivity improvements were observed in complex biological matrices, with SAMRS-based detection identifying B. cereus in milk at 100 CFU/mL versus 400 CFU/mL with conventional methods.
Enhancement of SNP Discrimination Capabilities
SAMRS technology significantly improves SNP discrimination by enhancing the specificity of primer-template interactions, particularly when combined with advanced detection platforms.
Table 2: SNP Discrimination Performance Across Different Technological Platforms
| Technology Platform | Principle | Discrimination Performance | Key Advantage |
|---|---|---|---|
| SAMRS-Enhanced Detection | Modified hydrogen bonding prevents mismatched hybridization | Near-absolute discrimination of single-base mismatches | Eliminates false positives from primer self-interaction |
| Cas12a with Strand Displacement | Cascade amplification with CRISPR activation | 0.1% variant allele frequency detection | PAM-independent recognition [90] |
| SWAT Method | Temperature-controlled washing based on mismatch-induced Tm reduction | >160:1 discrimination ratio | Adaptable to various biosensor platforms [91] |
The integration of SAMRS with isothermal amplification methods has demonstrated particular utility in SNP detection, achieving reliable discrimination of 0.1% single-base variations in biological samples including human buccal swabs [90]. This level of precision enables applications in pharmacogenomics and personalized medicine where accurate SNP genotyping is critical for treatment decisions.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for SAMRS Implementation
| Reagent/Chemical | Function/Application | Specific Example |
|---|---|---|
| SAMRS Nucleoside Phosphoramidites | Primer synthesis with altered hydrogen bonding capacity | 3-nitropyrrole (A analog), 2-nitropyrrole (G analog) [88] |
| EvaGreen Dye | Intercalating dye for real-time monitoring of amplification | Enables visualization without complex probe design [89] |
| Cas12a Enzyme | CRISPR-associated nuclease for sequence-specific detection | Provides trans-cleavage activity upon target recognition [90] |
| Thioflavin T (ThT) | G-quadruplex-specific fluorescent dye | Label-free detection in CRISPR-based assays [90] |
| Recombinase Enzymes | Strand invasion for isothermal amplification | Core component of RAA and RPA assays [89] |
Methodological Workflows
The following diagrams illustrate key experimental workflows and conceptual frameworks for implementing SAMRS technology.
Discussion and Future Perspectives
The implementation of SAMRS technology addresses fundamental limitations in molecular diagnostics by reengineering the hydrogen bonding properties of oligonucleotide primers. The quantitative data presented in this review demonstrates that SAMRS achieves near-complete elimination of primer dimers while simultaneously enhancing the fidelity of SNP discrimination. This dual capability positions SAMRS as a transformative technology for applications requiring exceptional specificity, including clinical diagnostics, pharmacogenomics, and low-abundance mutation detection.
Future developments in SAMRS chemistry will likely focus on expanding the repertoire of modified nucleobases with enhanced discriminatory properties and improved enzymatic compatibility. The integration of SAMRS with emerging detection platforms, particularly miniaturized point-of-care devices and multiplexed array technologies, promises to further advance the capabilities of molecular diagnostics. Additionally, the application of SAMRS principles to therapeutic oligonucleotides may open new avenues for controlling off-target effects and improving therapeutic indices.
As the field continues to recognize the critical importance of hydrogen bonding control in nucleic acid diagnostics, SAMRS and related technologies are poised to become standard tools for researchers and clinicians seeking unprecedented accuracy in genetic analysis.
The polymerase chain reaction (PCR) represents an indispensable tool in modern molecular biology, yet its "endless ability to confound" becomes particularly evident in multiplexed formats where multiple DNA targets are amplified simultaneously [43]. The fundamental challenge stems from primer-primer interactions, where multiple primers present in the same reaction mixture can hybridize to each other rather than to their intended target sequences, forming unproductive primer dimers [43]. These artifacts consume precious reaction resources—including primers, polymerase, and nucleotides—while generating competing amplification products that reduce sensitivity and specificity [43]. This problem intensifies as the level of multiplexing increases, with cross-reacting primers often defeating analyses involving more than a dozen target amplicons [46].
At the heart of this challenge lies the molecular recognition governed by hydrogen bonding between standard nucleobases. In natural DNA systems, complementary bases form specific hydrogen-bonded pairs: adenine (A) with thymine (T) via two hydrogen bonds, and guanine (G) with cytosine (C) via three hydrogen bonds [18]. Unfortunately, these same predictable pairing rules enable unintended interactions between primers when their sequences contain complementary regions, facilitating the formation of primer-dimers that sabotage assay efficiency [43].
Self-Avoiding Molecular Recognition Systems (SAMRS) represent a innovative approach to this fundamental problem. SAMRS employs synthetic nucleobase analogs designed to form stable base pairs with their natural complements but exhibit significantly reduced pairing with other SAMRS components [46] [92]. This introduction of asymmetric recognition capabilities creates a molecular environment where primers interact efficiently with their DNA targets while avoiding problematic interactions with each other, thereby addressing a core limitation in multiplex PCR through rational design of hydrogen bonding patterns.
The Molecular Basis of SAMRS Technology
Hydrogen Bond Engineering in Nucleobase Design
The SAMRS approach fundamentally reengineers the hydrogen bonding patterns of standard nucleobases to create a recognition system with built-in directional preferences. The system employs specifically modified nucleobases: 2-aminopurine (A) as an adenine analog, 2-thiothymine (T) as a thymine analog, 2′-deoxyinosine (hypoxanthine, G) as a guanine analog, and N4-ethylcytosine (C) as a cytosine analog [46] [44]. These modifications create strategic imbalances in hydrogen bonding capacity:
- A*:T Pairing - 2-Aminopurine pairs with natural thymine using its bottom two hydrogen bonding units, forming a stable duplex with two hydrogen bonds comparable to a natural A:T pair [46].
- T*:A Pairing - 2-Thiothymine pairs with natural adenine slightly better than natural thymine itself due to improved minor groove solvation [46].
- G*:C Pairing - Hypoxanthine (found in inosine) pairs with natural cytosine using its top two hydrogen bonding units [46].
- C*:G Pairing - N4-Ethylcytosine pairs with natural guanine using its bottom two hydrogen bonding units [46].
Critically, the SAMRS:SAMRS pairs (A:T and G:C) form only one hydrogen bond in standard Watson-Crick geometry, providing minimal contribution to duplex stability [46]. This strategic redesign of molecular recognition creates the foundational property of SAMRS: primers incorporating these modified bases bind efficiently to natural DNA templates but show significantly reduced affinity for other SAMRS-containing primers [92].
Historical Development and Validation
The conceptual foundation for SAMRS emerged from earlier work on "pseudocomplementary" nucleotides. In 1996, Kutyavin and colleagues demonstrated that diaminopurine and 2-thiothymine would bind to thymine and adenine respectively, but diaminopurine would not bind to 2-thiothymine [46]. This pseudocomplementary approach was initially employed in peptide nucleic acids (PNAs) to invade duplex DNA [46]. Subsequent work by Gamper showed that similar species as triphosphates could be incorporated into DNA, potentially producing products with reduced secondary structure formation for more uniform capture on arrays [46].
The development of a complete, functional SAMRS alphabet required extensive empirical optimization. Early candidates like zebularine derivatives as C* analogs proved chemically unstable under standard DNA synthesis conditions [46]. Similarly, while hypoxanthine (inosine) performed well as G* with some polymerases, many thermophilic DNA polymerases rejected it, possibly because it represents a deamination product of adenosine that occurs at high temperatures where extreme thermophiles live [46]. Through systematic evaluation, the current SAMRS alphabet emerged as the optimal combination for PCR applications, with N4-ethylcytosine selected over N4-methylcytosine due to its superior ability to distinguish between matched and mismatched pairs [46].
Table 1: SAMRS Components and Their Hydrogen Bonding Properties
| SAMRS Component | Natural Complement | H-Bonds with Natural Complement | SAMRS Complement | H-Bonds with SAMRS Complement |
|---|---|---|---|---|
| 2-Aminopurine (A*) | T | 2 | 2-Thiothymine (T*) | 1 |
| 2-Thiothymine (T*) | A | 2 (enhanced stability) | 2-Aminopurine (A*) | 1 |
| Hypoxanthine (G*) | C | 2 | N4-Ethylcytosine (C*) | 1 |
| N4-Ethylcytosine (C*) | G | 2 | Hypoxanthine (G*) | 1 |
Experimental Assessment of SAMRS Performance
Primer Design and Optimization Strategies
Implementing SAMRS technology requires careful consideration of both the number and placement of modified nucleotides within primer sequences. Research indicates that primers should be at least 20 nucleotides long and typically contain 1-3 SAMRS modifications [92]. A chimeric architecture often proves most effective, with SAMRS components concentrated in the 3′-segment that determines binding specificity and natural nucleotides in the 5′-segment [46]. This design leverages the self-avoiding property where it matters most—at the 3′ end where primer-dimer formation initiates—while maintaining overall duplex stability.
Not all SAMRS components contribute equally to duplex stability. The relative destabilization follows the order: T* (least destabilizing), followed by A* and C, with G being the most destabilizing [92]. This hierarchy must be considered during primer design, particularly for G-rich sequences that already tend to have lower amplification efficiency [92]. Additionally, experimental evidence suggests that keeping the 3′-most base as natural DNA typically yields better results [92].
The development of heuristics for Tm adjustment represents another critical advancement for practical implementation. By analyzing sequences and correlating them with experimental Tm data, researchers have developed estimates for how different nearest-neighbor doublets containing SAMRS components affect melting temperatures [43]. This enables more accurate prediction of primer behavior during the design phase, reducing the extensive empirical optimization traditionally required for multiplex assays.
Quantitative Analysis of Binding Affinities
Thermal melting studies provide crucial quantitative data on the binding properties of SAMRS-containing oligonucleotides. When introduced individually into reference DNA duplexes, each SAMRS:standard pair contributes to duplex stability approximately as well as a natural A:T pair [46]. However, the critical distinction emerges in SAMRS:SAMRS interactions, which in every case contribute less stability than the corresponding SAMRS:standard pair [46].
The strategic placement of SAMRS modifications significantly impacts primer performance. Studies examining primers with different numbers of SAMRS components placed at strategic positions reveal that even limited incorporation (4-8 components in a 25-mer) can dramatically reduce primer-dimer formation while maintaining efficient target amplification [46] [43]. This represents a significant advantage over earlier approaches that attempted to create entirely alternative genetic systems, as the chimeric design balances the benefits of self-avoidance with the practical requirements of polymerase recognition and duplex stability.
Table 2: Performance Comparison of Standard vs. SAMRS-Modified Primers in PCR
| Primer Type | Architecture | Primer-Dimer Formation | Target Amplification Efficiency | SNP Discrimination |
|---|---|---|---|---|
| Standard DNA | Fully natural | High | Failed due to primer-dimer [46] | Moderate |
| Hybrid SAMRS/Standard | One standard primer, one with 4 SAMRS | Moderate | Inefficient [46] | Improved |
| Full SAMRS (Low Modification) | Both primers with 4 SAMRS components | Low | Efficient [46] | High |
| Full SAMRS (High Modification) | Both primers with 8 SAMRS components | Very Low | Efficient [46] | Very High |
Experimental Protocols for SAMRS Implementation
Oligonucleotide Synthesis and Purification
SAMRS-containing oligonucleotides are synthesized using standard phosphoramidite chemistry on instruments such as the ABI 394 or ABI 3900 [43]. All SAMRS phosphoramidites are commercially available from suppliers like Glen Research or ChemGenes [43]. No special changes are needed for coupling and deprotection compared with standard phosphoramidites, following the synthesizer manufacturer's recommendations for dmf-dG, Ac-dC, and Bz-dA, dT [43].
For research applications, SAMRS-containing oligonucleotides can be synthesized either DMT-on or DMT-off. The DMT-off oligonucleotides are deprotected in aqueous ammonium hydroxide (28-33% NH3 in water) at 55°C for 10-12 hours, then purified by ion-exchange HPLC using columns such as Dionex DNAPac PA-100 (22 × 250 mm) and desalted over SepPak cartridges [43]. Oligonucleotides synthesized DMT-on are deprotected with ammonia, followed by DMT removal using Glen-Pak Cartridges; if purity falls below 80% by analytical ion-exchange HPLC, further purification by preparative ion-exchange HPLC is recommended [43]. For diagnostic kits, all SAMRS-containing oligonucleotides should be synthesized via the DMT-off strategy and purified by ion-exchange HPLC to meet stringent purity standards (>85% or 90%) [43].
Melting Temperature Analysis
The melting temperatures (Tms) of duplexes containing SAMRS components are measured in PCR buffer compatible with the intended polymerase (e.g., 1 µM of each oligonucleotide, 10 mM Tris-HCl, 50 mM KCl, pH 8.3 at 25°C, with 1.5 mM or 5.0 mM MgCl2 for JumpStart Taq DNA polymerase) [43]. Sequences typically incorporate strategic placements of SAMRS components, such as:
- Set 1 sequence: 5′-GAG CTG AGG TCA GTG T n n n n C-3′
- Set 2 sequence: 5′-GAG CTG AGG TCA GTG N n a t n N-3′
- Set 3 sequence: 5′-GCT CGA ATT GCA CCC T n n n n C-3′
(Upper case letters indicate standard nucleobases; lower case bold letters indicate SAMRS components) [43]
Melting curves are typically visualized using fluorescent dyes like EvaGreen (0.5×) in instruments such as the Roche LightCycler 480 with a temperature profile that includes: (i) denaturing and annealing duplexes: 95°C for 3 min, cool to 40°C with melting-curve setting (10 acq/°C; ~4-5°C/min), heat again to 50°C and hold for 10 min; (ii) slowly denature duplexes from 50°C to 90°C with melting-curve setting (100 acq/°C; ~1°C/min) [43]. Each set of duplexes should be measured in triplicate, with standard:standard and SAMRS:standard duplexes run in parallel on the same multi-well plate to ensure comparable conditions [43].
Applications and Performance in Complex Reactions
Enhanced Multiplex PCR Performance
The application of SAMRS technology to multiplex PCR demonstrates remarkable improvements in assay complexity and reliability. In one compelling experiment, ten pairs of chimeric {16+8*+1} primers (16 natural nucleotides + 8 SAMRS nucleotides + 1 natural nucleotide at the 3′ end) were designed to target 14 cancer-relevant genes [46]. These primers were intentionally not optimized by computer programs to avoid PCR artifacts, yet single-plexed PCR succeeded with all SAMRS-containing primer pairs [46]. Control primers with analogous sequences built entirely from standard nucleotides showed inconsistent performance, with one specific amplicon (PTPN11) failing completely in singleplex reactions [46].
The mechanism for this improved performance lies in the dramatic reduction of primer-primer interactions. In a striking example, primer pairs targeting the Taq gene with perfect complementarity in their last nine nucleotides were tested with varying SAMRS incorporation [46]. Primer pairs built from standard nucleotides failed completely to yield the desired amplicon, producing only primer-dimer artifacts [46]. When both primers contained four or eight SAMRS components, however, PCR amplification efficiently produced only the desired amplicon, demonstrating the powerful "SAMRS effect" even with relatively short modified segments [46].
Improved SNP Discrimination
SAMRS-modified primers offer significant advantages for single nucleotide polymorphism (SNP) detection, a critical application in clinical diagnostics and personalized medicine. The technology enables greater SNP discrimination than conventional allele-specific PCR, with the additional benefit of avoiding primer-dimer artifacts [43]. This enhanced specificity stems from the reduced stability of mismatched pairs when SAMRS components are positioned strategically within the primer sequence.
The improved discrimination power is particularly valuable for applications in oncology, where detecting specific mutations can guide targeted therapies. With appropriately chosen polymerases, SAMRS-based approaches can achieve superior allele discrimination while maintaining robust amplification of the correct target [43]. This combination of sensitivity and specificity addresses a fundamental challenge in molecular diagnostics, where false positives or negatives can have significant clinical consequences.
Compatibility with Polymerase Systems
The effectiveness of SAMRS technology depends critically on polymerase compatibility. Screening of numerous thermophilic DNA polymerases revealed that many from extreme thermophiles inefficiently incorporate SAMRS components, possibly because modified bases like hypoxanthine are recognized as deamination products [46]. However, Taq DNA polymerase performs well reading through SAMRS components in a template [46], making it a suitable choice for SAMRS-based assays.
Polymerase selection becomes particularly important when SAMRS components are incorporated near the 3′ end of primers, where extension efficiency is most critical. Studies have shown that 25-mer primers forming duplexes joined uniformly by two hydrogen bonds (characteristic of SAMRS:standard pairing) perform unpredictably, even at low temperatures with the Klenow fragment of DNA polymerase I [46]. This priming inefficiency correlates with the lower Tms of SAMRS:standard duplexes compared to standard DNA duplexes of the same length [46].
Research Reagent Solutions for SAMRS Applications
Successful implementation of SAMRS technology requires access to specialized reagents and tools. The following table outlines key resources for developing and optimizing SAMRS-based assays:
Table 3: Essential Research Reagents for SAMRS Experiments
| Reagent/Tool | Function | Examples/Suppliers |
|---|---|---|
| SAMRS Phosphoramidites | Chemical building blocks for oligonucleotide synthesis | Glen Research, ChemGenes [43] |
| DNA Synthesizers | Solid-phase oligonucleotide synthesis | ABI 394, ABI 3900 [43] |
| HPLC Purification Systems | Purification of SAMRS-containing oligonucleotides | Dionex DNAPac PA-100 columns [43] |
| Compatible DNA Polymerases | Enzymatic amplification of SAMRS-containing primers | Taq DNA polymerase [46] |
| Melting Temperature Instruments | Characterization of duplex stability | Roche LightCycler 480 [43] |
| Fluorescent Detection Dyes | Visualization of melting curves and amplification | EvaGreen [43] |
SAMRS technology represents a paradigm shift in addressing the fundamental challenge of primer-dimer formation in multiplex PCR. By strategically reengineering the hydrogen bonding patterns of nucleobases, SAMRS creates an asymmetric recognition system where primers maintain strong affinity for their natural DNA targets while avoiding problematic interactions with each other. The experimental evidence demonstrates that even limited incorporation of SAMRS components in chimeric primer designs can dramatically reduce artifacts while maintaining efficient target amplification across complex multiplex reactions.
The implications of this technology extend across diverse applications in clinical diagnostics, biomedical research, and biotechnology. As multiplexed analyses become increasingly central to personalized medicine, pathogen detection, and genetic testing, SAMRS offers a robust molecular solution to the scalability limitations that have traditionally constrained PCR-based assays. The continued refinement of SAMRS components, along with improved understanding of polymerase compatibility and primer design principles, promises to further expand the frontiers of multiplex molecular analysis.
Visual Guide: SAMRS Experimental Workflow
The following diagram illustrates the key stages in designing and implementing SAMRS-modified primers for multiplex PCR applications:
The formation of non-specific primer dimers during polymerase chain reaction (PCR) is a pervasive challenge in molecular diagnostics, leading to reduced assay sensitivity, specificity, and efficiency. This guide articulates a novel framework for diagnosing and mitigating primer dimer artifacts by applying the hydrogen-bond pairing principle, a concept refined through modern drug design. The core thesis posits that the strategic engineering of primer-terminal hydrogen-bond donor/acceptor pairs to create thermodynamically disfavored interactions with bulk water can significantly reduce non-specific hybridization. Supported by quantitative data, detailed protocols, and computational tools, this whitepaper provides diagnostic scientists with a rational methodology to design more accurate and robust molecular assays.
In molecular diagnostics, the reliability of nucleic acid amplification techniques is paramount. Primer dimers—spurious amplification artifacts formed by the cross-hybridization of primers—represent a significant source of false-positive results and reduced yield. The prevailing model attributes this primarily to transient, sequence-mediated base complementarity. However, this model provides an incomplete picture, overlooking the critical role of the aqueous solvent in which these interactions occur.
Concurrently, in pharmaceutical research, a persistent challenge has been the poor correlation between the strength of an individual hydrogen bond in a protein-ligand complex and the experimental binding affinity. The resolution to this puzzle emerged with the understanding that hydrogen bonds in biological systems do not form in isolation but in continuous competition with bulk water [93]. This led to the formulation of the hydrogen-bond pairing principle: a hydrogen bond enhances molecular interactions only when both the donor and acceptor have either significantly stronger or significantly weaker H-bonding capabilities than the hydrogen and oxygen atoms in water. Conversely, mixed strong-weak pairings decrease binding affinity due to competitive interference from water [93] [94].
This technical guide transposes this foundational principle from drug design to the realm of molecular diagnostics. It provides a new mechanistic explanation for primer dimer formation, framing it as a failure to manage the competitive H-bonding landscape with the solvent. The subsequent sections detail the quantitative principles, experimental methodologies, and computational tools required to systematically apply this knowledge to diagnostic assay design.
Core Principle: The Hydrogen-Bond Pairing Principle
Theoretical Foundation and Energetics
The fundamental reaction governing hydrogen bonding in an aqueous environment involves the exchange of H-bond partners between the receptor-ligand complex and the surrounding water molecules [93]. The free energy change (ΔG) for this competitive process determines whether a specific H-bond will form or be broken.
The H-bond pairing principle provides a predictive framework for this ΔG. The H-bonding capability of an atom is quantified by the free energy required to transfer it from water to a non-polar solvent like hexadecane, expressed as a ΔlogP₁₆ value [93]. Experimental data for key functional groups relevant to nucleic acids are summarized in Table 1.
Table 1: H-Bonding Capabilities of Nucleotide Functional Groups
| Atom/Functional Group | Context | H-bonding Capability (kJ/mol) | Comparison to H₂O |
|---|---|---|---|
| H₂O | - | 7.02 (±0.11) | Reference |
| Carbonyl O (Acceptor) | DNA/RNA Base | ~8.5 - 9.0 | Stronger |
| Amino N (Donor) | DNA/RNA Base | ~7.2 - 7.6 | Stronger |
| Hydroxyl O (Acceptor) | Sugar backbone | ~8.1 - 8.7 | Stronger |
| Apolar Atom | e.g., Carbon | 0 | Weaker |
The principle states:
- Synergistic Pairing: An H-bond enhances binding when donor and acceptor are both stronger or both weaker than water. This pairing minimizes the energetic penalty of desolvation, leading to a net gain in binding affinity.
- Antagonistic Pairing: An H-bond between a strong donor and a weak acceptor (or vice versa) decreases overall binding affinity. This mismatch creates a thermodynamic penalty as both atoms are forced to relinquish their optimal water interactions for a suboptimal partnership [93].
In the context of primer dimers, non-specific annealing can be driven by local, antagonistic H-bond pairings that are sufficiently stable in the absence of the true template. The strategic introduction of terminal bases that create antagonistic pairings can therefore destabilize these spurious interactions.
Visualizing the Competitive H-Bonding Landscape
The following diagram illustrates the competitive H-bonding process that determines the stability of a molecular interaction in an aqueous solution, as is the case for primer-template binding.
Diagnostic Application: Mitigating Primer Dimers
A Revised Model for Primer Dimer Formation
Traditional primer dimer models focus on Watson-Crick base pairing. The H-bond pairing principle adds a critical layer: the desolvation energy of the terminal bases. The initial docking of two primer ends is governed by the net free energy change of displacing water molecules from the potential H-bonding sites to form new inter-primer bonds.
Primer dimer formation is thus promoted when terminal nucleotides form multiple synergistic H-bond pairings, even with limited geometric complementarity. The energy gain from these pairings can be sufficient to overcome the entropic cost of the primers associating.
Practical Design Rules for Diagnostic Primers
Applying this principle leads to concrete design strategies:
- Terminal Mismatch Engineering: Deliberately design the 3'-end sequence so that any potential dimer partner for another primer is forced into antagonistic H-bond pairings. For example, if a terminal base has a strong H-bond donor, its partner in a potential dimer should be a weak acceptor.
- H-Bond Capability Analysis: During in silico primer design, screen not only for sequence complementarity but also for the summed H-bonding capability of potential dimer interfaces. Favor primer pairs whose terminal regions have low potential for forming strong-strong H-bond pairs.
- Exploiting Weak-Weak Pairings: Incorporate modified nucleotides or terminal bases that present H-bond donors/acceptors with capabilities weaker than water (e.g., certain halogenated bases) to create terminal "caps" that resist all non-specific H-bonding.
Table 2: Experimental Binding Affinity Changes from H-Bond Engineering
| Modification Type | H-Bond Pairing Created | Theoretical ΔΔG | Experimental Effect | Source Context |
|---|---|---|---|---|
| Single atom change in InsP6 | Strong-Strong | Favorable | 26-fold increased binding affinity to target [94] | Drug Design |
| Mixed Strong-Weak pairing | Strong-Weak | Highly Unfavorable | Up to 3-million-fold decrease in binding [94] | Drug Design |
| Theoretical for Primer: dG (3') to dC (3') | Strong-Strong | Favorable | Predicted to promote dimerization | This Guide |
| Theoretical for Primer: Modified Base (3') | Weak-Weak | Neutral/Favorable | Predicted to inhibit dimerization | This Guide |
Experimental Protocols and Workflows
Protocol 1:In SilicoH-Bond Topology Analysis of Primer-Primer Interactions
This protocol uses a bioinformatic approach to classify and predict the dimerization propensity of primer pairs based on their H-bond topology.
1. Input Preparation:
- Generate PDB-style coordinate files for all primer sequences in the assay. Software like Avogadro or PyMOL can be used for initial structure building.
- Ensure all hydrogen atoms are added and positioned correctly. Use a tool like MolProbity's
reduceto model hydrogen positions at biological pH uniformly [95].
2. Hydrogen Bond Identification:
- Use a geometric algorithm (e.g., a custom Python script or tools like HBPlus) to identify all potential hydrogen bonds [95].
- Criteria: Donor-Acceptor distance < 3.9 Å; Hydrogen-Acceptor distance < 2.5 Å; Donor-Hydrogen-Acceptor angle > 90°; Hydrogen-Acceptor-Antecedent angle > 90°.
3. Molecular Graph Construction:
- Represent the primer structure as a molecular graph, G=(V,E).
- Nodes (V): All atoms that are hydrogen bond donors and/or acceptors.
- Edges (E): Hydrogen bonds between donor and acceptor atoms that meet the geometric criteria [95].
4. Dimer Propensity Classification:
- Analyze the molecular graphs of potential primer dimers using a graph convolutional network (GCN) like HBcompare [95].
- The GCN will learn an embedding for each primer's H-bond topology and classify its likelihood of forming stable dimers with other primers in the set based on historical data of dimerizing vs. non-dimerizing pairs.
Protocol 2: Empirical Validation of Dimer Formation Using Melt Curve Analysis
1. Assay Setup:
- Prepare reaction mixtures containing the primer pair of interest, intercalating dye (e.g., SYBR Green I), buffer, and dNTPs, but no DNA template.
- Run the reaction on a real-time PCR instrument with a standard amplification cycle, followed by a high-resolution melt step from 60°C to 95°C.
2. Data Analysis:
- Plot the negative derivative of fluorescence with respect to temperature (-dF/dT vs. T).
- A low-temperature melt peak (e.g., 60-75°C) is indicative of primer dimer formation.
- Compare the melt curves and peak temperatures for primers designed with standard rules versus those engineered with the H-bond pairing principle.
3. Interpretation:
- A significant reduction in the height or area of the low-temperature melt peak, or its complete disappearance, indicates successful suppression of primer dimers through H-bond engineering.
Workflow for Primer Design and Validation
The following diagram outlines the integrated workflow from initial design to empirical validation.
Table 3: Key Research Reagent Solutions for H-Bond Studies
| Tool / Reagent | Function / Description | Application in This Field |
|---|---|---|
| HBondFinder | A computational tool that uses geometric criteria to identify all hydrogen bonds in a protein or nucleic acid structure from its PDB file [95]. | Generating molecular graphs of primers and potential dimers for topology analysis. |
| HBcompare | A deep learning algorithm that classifies protein structures by ligand binding preference based solely on hydrogen bond topology [95]. | Can be adapted to classify primer sequences by their dimerization propensity. |
| Graph Convolutional Network (GCN) | A type of neural network that operates directly on graph structures, ideal for learning from molecular graphs [95]. | The core engine of HBcompare for learning H-bond topology patterns. |
| COSMO-based Sigma Profiles | Quantum-chemically derived molecular descriptors that quantify a molecule's polarity and H-bonding character (acidity α, basicity β) [96]. | Predicting H-bond interaction energies for novel or modified nucleotides during the design phase. |
| Modified Nucleotides | Nucleotides with functional groups altering their inherent H-bonding capability (e.g., weaker than water). | Experimentally creating terminal "weak-weak" H-bond pairs to empirically test dimer suppression. |
The transposition of the hydrogen-bond pairing principle from drug design to molecular diagnostics provides a powerful, mechanistic framework for addressing the persistent challenge of primer dimers. By moving beyond a purely sequence-based view to one that incorporates the thermodynamics of competitive solvation, researchers can now rationally design primer termini that are inherently resistant to spurious annealing.
The experimental and computational protocols outlined herein offer a direct path to implementation. The future of this field lies in the expansion of quantitative H-bonding capability data for modified nucleotides, the tighter integration of tools like HBcompare into mainstream primer design software, and the application of these principles to other diagnostic artifacts, such as probe dimerization in multiplexed assays. By mastering the language of hydrogen bonds as spoken in an aqueous environment, the next generation of molecular diagnostics can achieve unprecedented levels of precision and reliability.
Conclusion
The formation of primer dimers is fundamentally a problem of unintended hydrogen bonding, a force that can be precisely understood and managed. By integrating foundational knowledge of biophysical chemistry with strategic primer design and rigorous troubleshooting protocols, researchers can effectively suppress these artifacts. The advent of novel technologies like SAMRS demonstrates a clear path forward, turning the challenge of hydrogen bonding into a design parameter that can be controlled. Future directions point toward the increased use of these sophisticated design rules and alternative chemistries to enable highly multiplexed, ultra-sensitive diagnostic assays with minimal background, thereby accelerating discoveries in biomedical research and improving the accuracy of clinical diagnostics. The principles of managing hydrogen bonding extend beyond PCR, offering valuable insights for general drug design and molecular recognition.
References
- 1. Troubleshooting primer dimer in PCR [https://www.minipcr.com/primer-dimer-pcr/]
- 2. Evidence for a Watson-Crick Hydrogen Bonding ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1190260/]
- 3. Primer Dimer - an overview [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/primer-dimer]
- 4. Guide to Primer Design for PCR [https://www.benchling.com/primer-design-for-pcr]
- 5. Impact of Primer Dimers and Self-Amplifying Hairpins on ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5922443/]
- 6. Definition, Types, Primer Design Online Tools, Uses [https://microbenotes.com/primer/]
- 7. Understanding qPCR Efficiency and Why It Exceeds 100% [https://biosistemika.com/blog/qpcr-efficiency-over-100/]
- 8. How to Design Primers for DNA Sequencing | Practical Lab ... [https://www.cd-genomics.com/resource-how-to-design-primers-for-dna-sequencing.html]
- 9. Prevention of pre-PCR mis-priming and primer ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC312262/]
- 10. Both aerosol and primer dimer breakdown for ... [https://www.sciencedirect.com/science/article/abs/pii/S0039914024016825]
- 11. Base pair [https://en.wikipedia.org/wiki/Base_pair]
- 12. Base pairs [https://www.genomicseducation.hee.nhs.uk/glossary/base-pairs/]
- 13. 20.20: The Double Helix [https://chem.libretexts.org/Bookshelves/General_Chemistry/ChemPRIME_(Moore_et_al.)/20%3A_Molecules_in_Living_Systems/20.20%3A_The_Double_Helix]
- 14. B-DNA structure and stability: the role of hydrogen bonding, π ... [https://pubs.rsc.org/en/content/articlehtml/2014/ob/c4ob00427b]
- 15. Complementary Base Pairing: Hydrogen Bonding [https://www2.chem.wisc.edu/deptfiles/genchem/netorial/modules/biomolecules/modules/dna1/dna15.htm]
- 16. Hydrogen Bonding in Natural and Unnatural Base Pairs—A ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8071019/]
- 17. PCR Primer Design Tips - Behind the Bench [https://www.thermofisher.com/blog/behindthebench/pcr-primer-design-tips/]
- 18. Primer design guide - 5 tips for best PCR results [https://the-dna-universe.com/2022/09/05/primer-design-guide-the-top-5-factors-to-consider-for-optimum-performance/]
- 19. Optimization of primer sets and detection protocols for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7295692/]
- 20. Archived | DNA Amplification | GC Content Percentage [https://nij.ojp.gov/nij-hosted-online-training-courses/dna-amplification/overview/primer-design/gc-content-percentage]
- 21. When designing primers how important is the GC clamp? [https://biology.stackexchange.com/questions/10843/when-designing-primers-how-important-is-the-gc-clamp]
- 22. PrimerMapper: high throughput primer design and ... [https://www.nature.com/articles/srep20631]
- 23. Primer designing tool - NCBI - NIH [https://www.ncbi.nlm.nih.gov/tools/primer-blast/]
- 24. URAdime – a tool for analyzing primer sequences in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11838186/]
- 25. How to design primers for PCR - INTEGRA Biosciences [https://www.integra-biosciences.com/united-states/en/blog/article/how-design-primers-pcr]
- 26. The importance of intramolecular hydrogen bonds on ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8693363/]
- 27. In Silico Design of Quantitative Polymerase Chain ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12025841/]
- 28. PCR Primer Design Guidelines [http://www.premierbiosoft.com/tech_notes/PCR_Primer_Design.html]
- 29. Mastering PCR Primer Design: The 12 Golden Rules for Su... [https://www.sbsgenetech.com/blog/mastering-pcr-primer-design-the-12-golden-rules-for-success?srsltid=AfmBOorTkarmOB8MKJoRwg_2QjsoHdA7plV9Xv69jNqaZ8cInSY4xGgv]
- 30. qPCR primer design revisited [https://www.sciencedirect.com/science/article/pii/S221475351730181X]
- 31. Primer Dimer - an overview [https://www.sciencedirect.com/topics/neuroscience/primer-dimer]
- 32. Covalent modification of primers improves PCR ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6994073/]
- 33. Rules and Tips for PCR & qPCR Primer Design | IDT [https://www.idtdna.com/page/support-and-education/decoded-plus/how-to-design-primers-and-probes-for-pcr-and-qpcr/]
- 34. 2025 Oligonucleotide Design Guide: Complete User Manual [https://oligopool.com/resources/user-guide]
- 35. Quantitating primer-template interactions using ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11317036/]
- 36. Dynamics of hydrogen-bonded end groups in bulk ... [https://www.nature.com/articles/s42004-025-01597-w]
- 37. A Computational Tool for Large-Scale Primer Design and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12469620/]
- 38. Model ISBICSKIS-BKK - iGEM 2025 [https://2025.igem.wiki/isbicskis-bkk/model]
- 39. Prediction of PCR amplification from primer and template ... [https://www.nature.com/articles/s41598-021-86357-1]
- 40. Inversing the natural hydrogen bonding rule to selectively ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2475633/]
- 41. or Fluorescent Probes For RT-qPCR? Intercalating Dyes [https://bitesizebio.com/36056/dyes-fluorescent-probes-rt-qpcr/]
- 42. Quantitative Experimental Determination of Primer-Dimer ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3416024/]
- 43. Eliminating primer dimers and improving SNP detection ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7200914/]
- 44. Self-Avoiding Molecular Recognition Systems (SAMRS) [https://pubmed.ncbi.nlm.nih.gov/18776287/]
- 45. Self-avoiding molecular recognition systems (SAMRS ... [https://www.biosyn.com/tew/Members-of-the-%E2%80%9CSelf-Avoiding-Molecular-Recognition-Systems%E2%80%9D-or-SAMRS-bind-to-natural-DNA-but-not-to-other-SAMRS-analogs.aspx]
- 46. Artificial Genetic Systems. Self-Avoiding DNA in PCR and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6027612/]
- 47. Self-avoiding molecular recognition systems in dna ... [https://patents.google.com/patent/EP2321332A1/en]
- 48. RNase H-dependent PCR (rhPCR) - Reduce Primer Dimers [https://eu.idtdna.com/pages/technology/qpcr-and-pcr/rhamp-pcr]
- 49. fever as a reemerging disease in upper Egypt: Diagnosis... Dengue [https://pmc.ncbi.nlm.nih.gov/articles/PMC9060354/]
- 50. Development and validation of RT-LAMP for detecting ... [https://www.nature.com/articles/s41598-024-74020-4]
- 51. Development and Validation of a Combined RT - LAMP for the... Assay [https://www.preprints.org/manuscript/202409.0463/v1]
- 52. Development of loop-mediated isothermal amplification ... [https://www.nature.com/articles/s41598-023-31760-z]
- 53. Development of a pan-serotype reverse transcription loop ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7126901/]
- 54. A quantitative RT‐qLAMP for the detection of SARS‐CoV‐2 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9349938/]
- 55. Reverse transcription loop-mediated isothermal ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10728416/]
- 56. Development of reverse-transcription loop-mediated isothermal... [https://bmcmicrobiol.biomedcentral.com/articles/10.1186/s12866-015-0595-1]
- 57. Analysis of a Reverse Transcription Loop-mediated ... [https://www.sciencedirect.com/science/article/pii/S0166093415003377]
- 58. Optimizing PCR: Proven Tips and Troubleshooting Tricks [https://www.the-scientist.com/optimizing-pcr-proven-tips-and-troubleshooting-tricks-71660]
- 59. Optimizing your PCR [https://www.takarabio.com/learning-centers/pcr/faq/optimization?srsltid=AfmBOopSjs1ZYSj8Dyqmz0Yq9ADTFvQQasSfwjQ-6BGce7omc5M-LwkY]
- 60. Touchdown PCR: Easy Intro Plus 5 Expert Tips [https://bitesizebio.com/2203/touchdown-pcr-a-primer-and-some-tips/]
- 61. Standard vs. Touchdown PCR [https://www.aatbio.com/resources/application-notes/standard-vs-touchdown-pcr]
- 62. Four tips for PCR amplification of GC-rich sequences [https://www.neb.com/en-us/nebinspired-blog/four-tips-for-pcr-amplification-of-gc-rich-sequences?srsltid=AfmBOorLEK621JVGazVeYmS_XG_EVWeXlr3Vd23PkjHYxBarpcHIdO0m]
- 63. Types, mechanisms, and applications in long-range PCR [https://www.sciencedirect.com/science/article/abs/pii/S0300908422000499]
- 64. Optimizing PCR Reaction Conditions for High Fidelity and ... [https://www.labx.com/resources/optimizing-pcr-reaction-conditions-for-high-fidelity-and-yield/5579]
- 65. Just What Do All These Additives Do? [https://bitesizebio.com/19420/just-what-do-all-these-additives-do/]
- 66. Platinum SuperFi II DNA Polymerase-High Fidelity PCR ... [https://www.thermofisher.com/us/en/home/life-science/pcr/pcr-enzymes-master-mixes/platinum-high-fidelity-pcr-enzyme.html]
- 67. Q5® High-Fidelity DNA Polymerase [https://www.neb.com/en-us/products/m0491-q5-high-fidelity-dna-polymerase?srsltid=AfmBOoqmlxIEWLWKUxIRENFNxwS26_Z9kH1sdYLiKDV35kK1xWTWWyT6]
- 68. High Fidelity, Long and GC Rich PCR - Molecular Biology [https://www.biotechrabbit.com/applications/molecular-biology-applications/high-fidelity-long-and-gc-rich-pcr.html]
- 69. DNA Polymerase Selection Chart [https://www.neb.com/en-us/tools-and-resources/selection-charts/dna-polymerase-selection-chart?srsltid=AfmBOorzFR6QdVA2ZFT7_q5lYHCsu4p8xf1hfLX8mNGyYsEs0-Mhm_Gg]
- 70. Selection Guide for Taq DNA Polymerase [https://www.promega.com/products/pcr/taq-polymerase/taq-dna-polymerase-selection-guide/]
- 71. Amplification of the No Template Control (NTC) [https://www.thermofisher.com/us/en/home/life-science/pcr/real-time-pcr/real-time-pcr-learning-center/real-time-pcr-basics/real-time-pcr-troubleshooting-tool/gene-expression-quantitation-troubleshooting/amplification-no-template-control.html]
- 72. Avoiding false positives with PCR | IDT [https://www.idtdna.com/page/support-and-education/decoded-plus/how-to-avoid-false-positives-in-pcr-and-what-to-do-if-you-get-them/]
- 73. High-Resolution Melt Analysis for Rapid Comparison of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4054126/]
- 74. Multiple Primer Analyzer | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/multiple-primer-analyzer.html]
- 75. Designing highly multiplex PCR primer sets with Simulated ... [https://www.nature.com/articles/s41467-022-29500-4]
- 76. Optimised genome editing for precise DNA insertion and ... [https://elifesciences.org/reviewed-preprints/107475]
- 77. High-throughput PRIME-editing screens identify functional ... [https://www.sciencedirect.com/science/article/pii/S1097276523009668]
- 78. Computational design of sequence-specific DNA-binding ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12618268/]
- 79. Requirement of Watson-Crick Hydrogen Bonding for DNA ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC162216/]
- 80. Comparison of different real-time PCR chemistries and their ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2322970/]
- 81. Comparison of different real-time PCR chemistries and their ... [https://bmcbiotechnol.biomedcentral.com/articles/10.1186/1472-6750-8-26]
- 82. qPCR and RT-qPCR [https://www.promega.com/resources/guides/nucleic-acid-analysis/qpcr-and-rt-qpcr-amplification/]
- 83. Comparison of Analytic Methods for Quantitative Real-Time ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4642823/]
- 84. Evaluation validation of a qPCR curve analysis method and ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-021-07986-4]
- 85. Tips for Designing PCR Primers [https://blog.edvotek.com/2025/08/14/tips-for-designing-pcr-primers/]
- 86. RNase H-dependent PCR enables highly specific ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0241803]
- 87. Polymerase Chain Reaction (PCR) - StatPearls - NCBI - NIH [https://www.ncbi.nlm.nih.gov/books/NBK589663/]
- 88. PCR Optimization: Strategies To Minimize Primer Dimer [https://genemod.net/blog/pcr-optimization]
- 89. Development of A Novel EvaGreen-Dye Based ... [https://weekly.chinacdc.cn/en/article/doi/10.46234/ccdcw2025.079]
- 90. Single nucleotide polymorphism discrimination and ... [https://www.sciencedirect.com/science/article/abs/pii/S0925400524015624]
- 91. A Novel Method That Allows SNP Discrimination with 160:1 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8391390/]
- 92. Glen Report 36-23: Application Note Polymerase Chain Reaction (PCR) Optimization [https://www.glenresearch.com/reports/gr36-23]
- 93. Regulation of protein-ligand binding affinity by hydrogen ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4820369/]
- 94. Hydrogen-bonding pairing helps design better drugs to ... [https://www.texaschildrens.org/content/news/hydrogen-bonding-pairing-helps-design-better-drugs-neutralize-gut-bacterial-infections]
- 95. HBcompare: Classifying Ligand Binding Preferences with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9687905/]
- 96. Prediction of hydrogen-bonding interaction energies with ... [https://www.sciencedirect.com/science/article/pii/S0167732225000650]